Bio Sensors and Food Protection
The potential threat of bioterrorism and emerging diseases requires advances in biosensor technologies to meet requirements for food protection in the 21st century.
Although concerns about the safety of food products have existed for a very long time, it is only recently that we have begun to take a national security–based interest in both the monitoring and protection of the food supply and distribution pipeline.

Food production, preparation, and sales are highly regulated in the United States and elsewhere around the world. This regulation is, in part, designed to ensure that food does not contain any of a number of naturally occurring pathogens. The enforcement of these regulations involves the implementation of safety procedures, good handling practices, and routine testing. However, relatively recent advances in micro and molecular biology, as well as the unfortunate increase in both actual and potential bioterrorism events, demand further evolution of the technology used for food protection and safety. The technology used in assuring food safety for the present and the future must address the new issues and capabilities that are a product of the world in which we live.
Few major incidences of intentional contamination of large food stocks occurred in the recent past. A 1984 incident in which a religious sect, the Rajneeshees, intentionally poisoned salad bars and coff ee creamers in the Oregon area with Salmonella was particularly notable (Carus, 2002). However, since the terrorist attacks of 2001, national concern about preventing and detecting bioterrorist attacks on food and food-related items has escalated. A number of incidences, including attacks by disgruntled employees of supermarkets, etc., have led to the realization that this type of terrorism is all too possible.
To deal with the threat of bioterrorist attack on the food supply, we will need to develop new capabilities in our food protection arsenal. Current methods for dealing with food safety include the ability to detect and identify organisms down to the serotype or serovar level. These methods are generally based on the assumption that the organisms of interest can be cultured and that the organism identity is among those that have been previously encountered and thoroughly characterized.
Increasing our capacity to protect the food supply from new pathogens and intentional contamination events is a complex issue. There is a need to consider the possibility that any pathogenic organism could be introduced into a food product, not just organisms known to be naturally present. Some of these organisms may be very difficult to culture, and there is the real possibility that the organisms could be genetically altered to make them appear diff erent, both physically and genetically, from any previously encountered pathogen.
Furthermore, in the case of intentional contamination, there is a need to increase our ability to provide a forensic analysis of the pathogen to aid in tracking both the source of the event and the spread. Current technologies for the detection of pathogens in food are not well suited for these types of problems.
Detecting and Identifying Pathogens
While the strict defi nition of a biosensor varies, it is generally considered to be an assay, product, or device that, by the use of a variety of methods, is capable of identifying biological material. This material ranges from chemical compounds, metabolites, proteins, and nucleic acids to entire living organisms.
The goal of using a biosensor in the role of food protection is to detect and identify organisms and biological products that could be considered harmful to end users of food products. The success of any biosensor will depend entirely on the technology used with the assay, product, or device.
--- PAGE BREAK ---
As discussed below, the current technologies available for testing food products for both naturally occurring and intentionally introduced pathogens are a mixture of methods and assays that have been available for many years, such as culture, biochemical analysis, and immunoassays. The vast majority of these assays were originally designed for basic microbiology and clinical applications and have been subsequently adopted for use in food product monitoring.
Several biosensors rely on these types of techniques (Eshkenazi et al., 2000; Geng et al., 2004; Taitt et al., 2004; Ricci and Palleschi, 2005; Sapsford et al., 2005). Recent developments of new, more powerful, assay types that depend on the ability to perform genome-level DNA sequencing and polymerase chain reaction (PCR)–based DNA reactions have added the ability to identify organisms somewhat more quickly and to genotype organisms at a much better level. New advances in biosensors have, and will continue to, incorporate these new technologies.
• Culture Methods. Perhaps the oldest method for the detection and identifi cation of food contaminants is culturing the sample of interest. Direct culturing of samples, often on selective media designed to enhance the growth of the expected organism, is followed by isolation of single colonies and subsequent morphological and biochemical analysis to assign a specifi c identification to the organism.
This method of detection has a number of distinct advantages, including ease of use, low cost, superb sensitivity, and the ability to rely on tremendous amounts of historical information. However, the method has some extreme limitations. The time to a high-confidence answer can range from 24 hr to several weeks, depending on the organism. Furthermore, viability of the sample becomes a major issue, since the organisms must grow in order to be identified, and the correct choice of selective media must be made. In addition, some knowledge of the biochemical indicators of the organism’s identity must be available.
In short, culture and biochemical analysis can be very useful techniques, assuming that one knows something about the organisms of interest in any particular sample. However, for unknown organisms, mixtures of organisms, or organisms in which indicators may have been altered, these types of techniques can often not answer the questions of interest.
• Immunoassays. Immunologically based assays, such as enzyme-linked immunoassays (ELISAs), fl uorescence-based sandwich immunoassays, Western blots, agglutination assays, etc., have all been used in microbial detection and identifi cation and can all be used in food protection assays.
Many immunoassays are available as dipstick assays directed against specific organisms. These types of assays are specifi c, reliable, reproducible, and aff ordable. However, their sensitivity is variable and may be insufficient for some low-level detections. Other potential negatives to these types of assays include diffi culty of producing the antibodies required, a high frequency of non-specifi c binding and inhibition by high backgrounds, and the possible requirement for single organism isolation (utilizing culture, with all of its issues).
• DNA Methods. Culture, biochemical assays, and immunological methods have been a part of pathogen detection and characterization in food products for a long time. More recently, genomelevel sequencing and PCR-based methods have entered the picture.
The ability to sequence large portions of bacterial and viral genomes gives investigators the tools needed to assign much-more-specifi c identities to organisms of interest. Not only can genes of interest be readily identifi ed, but single-nucleotide polymorphisms (SNPs) as well as small insertions and deletions can be easily characterized. However, this type of analysis requires relatively large amounts of starting material from single colony or organism isolates, and, while it provides a great deal of information, sequence analysis can be time-consuming and labor-intensive.
--- PAGE BREAK ---
PCR-based methods involve the specific amplifi cation of regions of the microbial genome and the subsequent use of these amplified regions to detect, characterize, and identify the organisms from which they were derived. Traditional PCR can be used to amplify specific regions of a microbial pathogen, either from a cultured sample or directly from a food product (following purification of the genomic material). When used with a specific genomic target region, the very presence of a product from the PCR reaction (an amplicon) is in itself an indicator of the presence and identity of the organism.
While the PCR reactions themselves are quick (0.5–2.5 hr), analysis of the products can be diffi cult. Typically, PCR reactions are analyzed by gel electrophoresis, which can be used to directly visualize the PCR products and give a good approximation of their size and quantity. Further information can be obtained by sequencing the PCR product, although the time and expense involved in this is not trivial. Using a number of modifications of the traditional techniques, PCR products can also be analyzed in real-time (RT-PCR), using fl uorescencebased detection methods.
PCR methods are extremely attractive in that they are sensitive (often down to a single copy of a genome), potentially quantitative, fast, and able to be performed on complex samples. The assays are limited to an extent by the need to have specific information about the target organisms and by an inability to look at large numbers of organisms in complex mixture simultaneously.
The TIGER Biosensor
The existing methods discussed above have served to determine the presence and identity of pathogens in food products. However, with the increased concern about the ability of bioterrorists to aff ect the world’s food production, storage, and consumption, and the tremendous increase in the technology available to modify pathogens and produce large quantities of infectious agents, there is a need to advance beyond these tests and technologies.
It is the ability to modify pathogenic and nonpathogenic organisms and then introduce these modified organisms that leads to the conclusion that our current technologies are simply not suitable for the situation at hand. The new questions for the 21st century are how do we detect and identify something we did not expect or may never have seen before and how can we characterize organisms to the point that we can pinpoint the source of the contamination exactly and assign a bioterrorist source if need be. The food industry needs to able to answer these questions and to screen large numbers of samples quickly.
Addressing the above issues requires a fundamental change in the way in which organisms are detected and characterized. One such solution is being utilized in the design and implementation of the biosensor system called TIGER, for Triangulation Identifi cation for the Genetic Evaluation of Risks (Hofstadler et al. 2005). This system, a smaller version of which is shown in Figure 1, was initially developed by Isis Pharmaceuticals in collaboration with Science Applications International Corporation (SAIC) and Defense Advanced Research Projects Agency (DARPA), and was subsequently funded by the National Institutes of Health (NIH) and the Centers for Disease Control and Prevention (CDC) for applications involving epidemic surveillance and detection of emerging infectious diseases. The system employs a high-performance electrospray mass spectrometry time-of-flight (TOF) instrument to derive base compositions of PCR products.
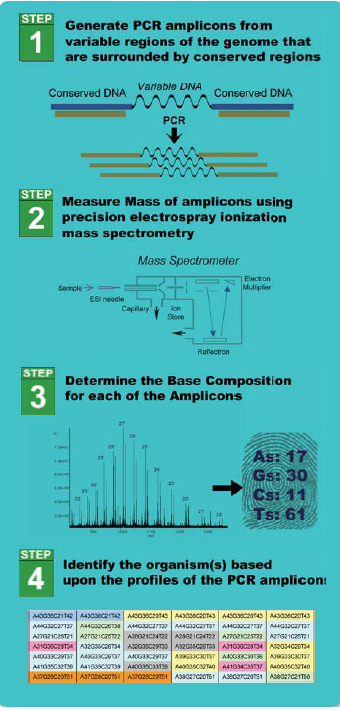 The operating principle of the system is shown in Figure 2. The process begins with the extraction of all nucleic acid present in a sample. The resulting nucleic acid material is divided up as input to a number of PCR reactions, each with a single broad-range primer pair used for the amplification reactions. The PCR reactions take place in a 96-microwell format, and the resulting amplicons are purifi ed for electrospray into a mass spectrometer.
The operating principle of the system is shown in Figure 2. The process begins with the extraction of all nucleic acid present in a sample. The resulting nucleic acid material is divided up as input to a number of PCR reactions, each with a single broad-range primer pair used for the amplification reactions. The PCR reactions take place in a 96-microwell format, and the resulting amplicons are purifi ed for electrospray into a mass spectrometer.
--- PAGE BREAK ---
The resulting spectral signals are processed to produce a list of all detected masses, and the resulting masses are then converted into an unambiguous base composition of adenosines, guanosines, cytidines, and thymidines. Using the information from all the primer pairs, the identities of the organisms in the sample can be deduced. Use of an internal calibrant in the samples allows for the determination of the quantity of input material used.
While this process depends on a PCR-based set of reactions, it is the unique set of primers and the unique form of the product analysis that really lends itself to changing the way in which this type of data can be used to detect and identify, down to a strain level, pathogens found in a food matrix.
The use of primer pairs that can each prime the production of amplicons from hundreds, if not thousands, of organisms removes the constraint of knowing exactly what one is looking for. Because the primers are, in most cases, designed against absolutely required and broadly conserved genetic elements, it is unlikely that these regions can be targeted by bioterrorists, as manipulation of these sites would render the organism nonviable. And the decoding of these amplicons into base compositions, while not actually providing exact sequence information, provides a very informative fi ngerprint. The use of the mass spectrometry instrument then allows the detection of all the amplicons produced simultaneously, without the need for pre-separation of the components.
There are a number of potential uses of the information provided by the use of broad primers and the ability to obtain base compositions from all of the amplicons produced in a complex reaction:
First is the obvious outcome of identifying a previously known organism. By matching base compositions from several primer pairs to those stored in a large database, an exact identification can be made.
Second is assigning identification to an unknown organism. This is made possible by the examination of the near-neighbor relationships of the base compositions of the resulting amplicons vs other known organisms (Sampath et al., 2005). Much like evolutionary trees created from sequence alignments, base compositions can also be used to fi nd closely related organisms and to assign a tentative identifi cation to an unknown organism (Ecker et al., 2005).
--- PAGE BREAK ---
Third is assigning a detailed strain identification or genotype to the organism. By using a diff erent set of primers, organisms can be organized by the fi ne diff erences in small regions of their genomes.
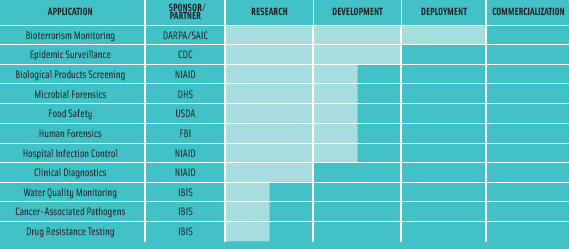 Other uses for the system beyond food safety also exist. These include human and animal disease diagnostics, human and animal forensics (Hall et al., 2005), and detection of adventitious organisms in drug products and vaccines. Development of these applications and others is currently underway in collaboration with a variety of partners and sponsors (Figure 3).
Other uses for the system beyond food safety also exist. These include human and animal disease diagnostics, human and animal forensics (Hall et al., 2005), and detection of adventitious organisms in drug products and vaccines. Development of these applications and others is currently underway in collaboration with a variety of partners and sponsors (Figure 3).
Fundamental Changes Ahead
It is very clear that the question is not whether we need biosensors for the purpose of food protection, but rather what format the assays contained within these biosensors need to take to be eff ective in the modern era.
It is no longer suffi cient for a biosensor to simply look for a previously characterized organism or, indeed, to look for any one type of organism. No longer can we predict what pathogen may be found in a food product or what exact form the pathogen might take. It is also clear that we have to move beyond the scope of random testing of samples of food products and more toward the testing of larger and larger numbers of samples to create as small an opportunity as possible for bioterrorists, and nature, to contaminate our food supply.
These needs require that future biosensors be fast, effi cient, readily available, and relatively inexpensive to use. No single biosensor currently meets all the needs described above, but newer biosensor systems, such as TIGER, are a solution to many of these problems.
One thing is clear—technological needs have always been met with exciting new types of inventions. The near future holds great promise for fundamental changes in our sensor and screening technology, and the result will be an increase in food safety around the world.
by Lawrence B. Blyn
is Executive Director of Biology, Ibis Div.,
Isis Pharmaceuticals, 1891 S. Rutherford Rd., Carlsbad, CA 92008
([email protected]).
References
Carus, W.S. 2002. “Bioterrorism and Biocrimes: The Illicit Use of Biological Agents Since 1900.” Fredonia Books, Amsterdam, The Netherlands.
Ecker, D.J., Sampath, R., Blyn, L.B., Eshoo, M.W., Ivy, C., Ecker, J.A., Libby, B., Samant, V., Sannes-Lowery, K., Melton, R.E., Russell, K., Freed, N., Barrozo, C., Wu, J., Rudnick, K., Desai, A., Moradi, E., Knize, D.J., Robbins, D.W., Hannis, J.C., Harrell, P.M., Massire, C., Hall, T.A., Jiang, Y., Ranken, R., Drader, J.J., White, N., McNeil, J.A., Crooke, S.T., and Hofstadler, S.A. 2005. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc. Natl. Acad. Sci.USA 102: 8012-8017.
Eshkenazi, I., Maltz, E., Zion, B., and Rishpon, J. 2000. A three-cascaded-enzymes biosensor to determine lactose concentration in raw milk. J. Dairy Sci. 83: 1939-1945.
Geng, T., Morgan, M.T., and Bhunia, A.K. 2004. Detection of low levels of Listeria monocytogenes cells by using a fi ber-optic immunosensor.” Appl. Environ.Microbiol. 70: 6138-6146.
Hall, T.A., Budowle, B., Jiang, Y., Blyn, L., Eshoo, M., Sannes-Lowery, K.A., Sampath, R., Drader, J.J., Hannis, J.C., Harrell, P., Samant, V., White, N., Ecker, D.J., and Hofstadler, S.A. 2005. Base composition analysis of human mitochondrial DNA using electrospray ionization mass spectrometry: A novel tool for the identifi cation and diff erentiation of humans.” Anal. Biochem. 344(1): 53-69.
Hofstadler, S.A., Sampath, R., Blyn, L.B., Eshoo, M.W., Hall, T.A., Jiang, Y., Drader, J.J., Hannis, J.C., Sannes-Lowery, K.A., Cummins, L.L., Libby, B., Walcott, D.J., Schink, A., Massire, C., Ranken, R., White, N., Samant, V., McNeil, J.A., Knize, D., Robbins, D., Rudnik, K., Desai, A., Moradi, E., and Ecker, D.J. 2005. TIGER: The universal biosensor. Intl. J. Mass Spectrom. 242: 23-41.
Ricci, F. and Palleschi, G. 2005. Sensor and biosensor preparation, optimisation and applications of Prussian Blue modifi ed electrodes. Biosens. Bioelectron. 21: 389-407. Epub Jan. 13.
Sampath, R., Hofstadler, S.A., Blyn, L., Eshoo, M., Hall, T., Massire, C., Levene, H., Hannis, J., Harrell, P.M., Neuman, B., Buchmeier, M.J., Jiang, Y., Ranken, R., Drader, J., Samant, V., Griff ey, R.H., McNeil, J.A., Crooke, S.T., and Ecker, D.J. 2005. Rapid identifi cation of emerging pathogens: Coronavirus. Emerg. Infect. Dis. 11: 373-379.
Sapsford, K.E., Taitt, C.R., Loo, N., and Ligler, F.S. 2005. Biosensor detection of Botulinum toxoid A and staphylococcal enterotoxin B in food. Appl. Environ. Microbiol. 71: 5590-5592.
Taitt, C.R., Shubin, Y.S., Angel, R., and Ligler, F.S. 2004. Detection of Salmonella enterica serovar typhimurium by using a rapid, array-based immunosensor. Appl. Environ. Microbiol. 70: 152-158.
