Big Projects Targeting Tiny Pathogens
FOOD SAFETY & QUALITY
The traditional method for identifying foodborne pathogens involves sampling and time-intensive culturing. Over the past decade or so, advancements in DNA technology such as whole genome sequencing (WGS) have led to more rapid and precise results, and a number of large-scale projects are using it to identify and trace foodborne pathogens.
 Sequencing the Whole Genome
Sequencing the Whole Genome
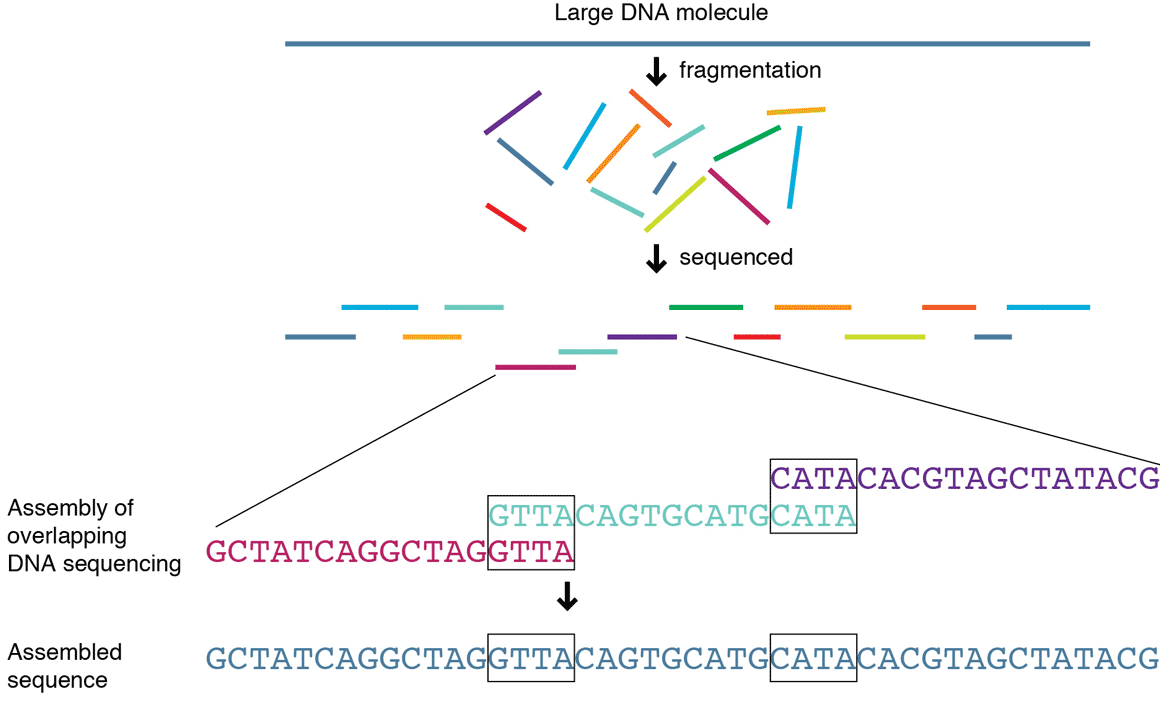
The sequence of bases (A, C, G, and T) in a DNA molecule carries the information a cell needs to assemble protein and RNA molecules. WGS is a process for determining the complete DNA makeup, or genome, of an organism, using specialized instrumentation and massive amounts of data processing. The process, which can be used to map genomes of known and unknown organisms, involves breaking the organism’s long DNA strands into shorter fragments called reads, which are sequenced individually. A computer program, or algorithm, looks for overlaps in the sequences and aligns the reads in their correct order to reconstitute the genome. The resulting genome can be compared to a reference genome to detect genetic variations. Next-generation sequencing (NGS), also known as high-throughput sequencing, is a WGS method that offers much more data at a given cost than previous sequencing methods and is increasingly being used by the technical community.
WGS is becoming less expensive and easier to use; has identical sample preparation for all pathogens; is the most accurate and high-resolution subtyping technique; and yields information about resistance, serotype, virulence factors, and other information in a single test. It can be used to monitor ingredient supplies, determine the effectiveness of preventive and sanitary controls, develop new rapid analytical methods, determine the persistence of pathogens in the environment, understand how pathogens spread within and between geographic areas and the effect of growing conditions and practices, and monitor emerging pathogens.
In addition, WGS can differentiate virtually all strains of foodborne pathogens, regardless of species. And because the genomic information of a species of foodborne pathogen found in one geographic area differs from that of the same species found in another area, it can be a powerful tool in tracking down the original source of contamination for a food product, especially if its ingredients come from different states or countries. It is also being used to study antimicrobial resistance.
Studying the Food Supply Chain
IBM, Armonk, N.Y. (ibm.com); Mars Inc., McLean, Va. (mars.com); and Bio-Rad Laboratories Inc., Hercules, Calif. (bio-rad.com), are conducting what they call the largest-ever metagenomics study to sequence the DNA and RNA of major food ingredients in various environments, at all stages in the supply chain, to help prevent foodborne illness and identify outbreaks before they begin. IBM and Mars established the Consortium for Sequencing the Food Supply Chain in January 2015, and Bio-Rad joined the consortium in January 2016. David Chambliss, lead researcher, food safety, at IBM Research–Almaden in San Jose, Calif., said that discussions are underway with other companies and academic institutions to join the consortium.
The three companies are participating in the scientific experimentation, interpretation of data, and project planning, Chambliss said. Mars provides microbiome samples and expertise in food processing and food safety management; with its expertise in computational systems and methods, IBM provides efficient processing of sequence data into usable knowledge; and Bio-Rad provides expertise in chemical analytics and sample preparation methods.
The initial technical work is being conducted at Mars’s pet food factory near Reno, Nev.; at the University of California, Davis; and at IBM Research–Almaden. It is focused on understanding the microbes present in the ingredients used in pet food manufacturing (such as poultry meal, meat and bone meal, fish meal, maize, and maize gluten) and analyzing what the microbial community can tell about unexpected things that might be in the ingredients. The work also involves validating the testing methods for increased speed and accuracy.
Samples are drawn from each shipment of a given ingredient on arrival at the factory, consolidated from multiple locations within the shipment, which for some ingredients may yield many samples per week. Selected samples are prepared for sequencing by extracting RNA and converting it to complementary DNA, and these prepared solutions, called libraries, are sent to a sequencing center. In some cases, microbial DNA is also extracted and sequenced. The raw RNA sequence data are analyzed to determine microbial composition and the relative expression of different microbial genes (an indication of biological activity). In the exploratory phase, sequencing and analysis are batched up periodically.
The project focuses on sequencing RNA rather than DNA, Chambliss said, and the analysis is based mainly on high-throughput, short-read technology, which measures the nucleotide sequences in hundreds of millions of molecules simultaneously to yield vast amounts of data in relatively few hours. Many instruments and kits from leading suppliers are used for sample preparation, library preparation, characterization, and additional testing. WGS is part of the testing, he said, but because RNA is being sequenced, what is really being done is whole transcriptome sequencing. Whole RNA sequencing gives exposure to all genes in the microbiome, not just a selected few as used in, for example, 16S amplicon sequencing. The use of whole genomes or transcriptomes is essential to learn about hazards and hazard responses, he said.
 Regarding whether the experiments have to be run on every ingredient or product that a company makes, Chambliss said that microbiome characterization will yield insights of compelling value across pretty much all ingredients, products, and even food categories, and the same core technologies of sequencing and informatics have very broad applicability. But the value of better knowledge about one ingredient or product can stand on its own without requiring that everything be studied equivalently. Each new ingredient or product, he said, has its own characteristics that call for study and analysis of how to generate and use genomic/transcriptomic results. The different fats, proteins, and enzymes in different foods call for variations in sample preparation. A biological genus that may routinely appear in fish meal might be a surprising indicator when found in poultry meal and even alarming if found on lettuce headed to consumers, he said.
Regarding whether the experiments have to be run on every ingredient or product that a company makes, Chambliss said that microbiome characterization will yield insights of compelling value across pretty much all ingredients, products, and even food categories, and the same core technologies of sequencing and informatics have very broad applicability. But the value of better knowledge about one ingredient or product can stand on its own without requiring that everything be studied equivalently. Each new ingredient or product, he said, has its own characteristics that call for study and analysis of how to generate and use genomic/transcriptomic results. The different fats, proteins, and enzymes in different foods call for variations in sample preparation. A biological genus that may routinely appear in fish meal might be a surprising indicator when found in poultry meal and even alarming if found on lettuce headed to consumers, he said.
At this point, the researchers are not narrowing down the potential uses of the data obtained. They often find new analyses to perform to draw new insights from the data. The consortium has developed an informatics platform designed to foster free-range exploration. A main goal, however, is to understand the relationship between microbial communities and the hazards that might be present in the food. While the potential hazards include well-known pathogenic organisms, he said, they also extend to non-microbial hazards to which the extant microbes respond in their growth and biological activity. So far, Chambliss said, the researchers have sequenced data from a range of ingredients and from a good number of shipments of one ingredient and are preparing papers for submission to refereed scientific journals to present the microbiological insights they are finding in the similarities and differences among those samples.
--- PAGE BREAK ---
Using Bioinformatics
Noblis, Falls Church, Va. (noblis.org), has developed a whole genome analysis tool called BioVelocity that provides insight into the molecular mechanisms of pathogen evolution, lineage calculations, and the emergence of highly pathogenic strains and is working with food industry partners in a pilot program to investigate ways of using this technology to enhance their food safety operations.
Using high-performance computing, BioVelocity identifies single nucleotide polymorphisms (SNPs) (variations in a single base pair of a DNA sequence) from NGS reads. The algorithm uses the reads as input, aligns the sequences simultaneously, and compares the results to a collection of reference genomes from the National Institutes of Health’s National Center for Biotechnology Information (NCBI) and Noblis’s own database of customized references to determine strain-level identification of foodborne pathogens in minutes.
Mitchell Holland, lead bioinformatician at Noblis, said that the pilot program is focused on applying WGS to identify pathogens and trace their sources during foodborne illness outbreaks in a timely manner. The food industry in general has not implemented this technology yet, he said, but continues to focus on using traditional methodologies to identify and control pathogens. The U.S. Food and Drug Administration (FDA) has been sequencing pathogens from foodborne outbreaks, and Noblis hopes to bridge the gap between industry and government agencies.
Noblis enlisted four partners in the pilot program to explore the potential of WGS: two food companies, one testing laboratory, and one academic institution conducting research on source tracing. The partners send samples to a third-party laboratory for NGS. Noblis then runs the sequence data through BioVelocity and provides the resulting identifications and phylogenetic information to the partners, with the hope that they will see the benefit of WGS over traditional microbiological assays. The company’s goal, Holland said, is to present a different perspective on microbial identification.
Network Provides Genome Database
The FDA has been utilizing WGS since 2008 and has been coordinating efforts by federal, state, and international public health agencies to sequence pathogens collected from foodborne outbreaks, contaminated food products, and environmental sources and make their genomic sequences publicly available in a database called Genome Trakr, which is housed at the NCBI. Using WGS for pathogen identification, laboratories in the Genome Trakr network upload their sequencing results to the database, and global companies conducting WGS on bacteria isolated from their sampling efforts can compare their results to the genomic information available in the database.
 Marc Allard, director of the FDA’s WGS laboratory, said that the network consists of 14 federal laboratories, 14 state health and university laboratories, one U.S. hospital laboratory, nine laboratories located outside the United States, and collaborations with independent academic researchers. The network has sequenced more than 57,000 isolates to date and completed more than 100 genomes. The network regularly sequences more than 1,000 isolates each month. The laboratories initially focused their WGS efforts on Salmonella and Listeria but have expanded their efforts to Escherichia coli, Campylobacter, Vibrio, Cronobacter, and other isolates as well as parasites and viruses.
Marc Allard, director of the FDA’s WGS laboratory, said that the network consists of 14 federal laboratories, 14 state health and university laboratories, one U.S. hospital laboratory, nine laboratories located outside the United States, and collaborations with independent academic researchers. The network has sequenced more than 57,000 isolates to date and completed more than 100 genomes. The network regularly sequences more than 1,000 isolates each month. The laboratories initially focused their WGS efforts on Salmonella and Listeria but have expanded their efforts to Escherichia coli, Campylobacter, Vibrio, Cronobacter, and other isolates as well as parasites and viruses.
GenomeTrakr continues to grow and expand with new isolates as well as new partner states, and most new growth will come from industry collaboration and international partners, Allard said. This new tool for epidemiologists helps solve outbreaks faster and identifies the root cause of contamination events to help develop preventive controls for broader food safety, he added.
Other Activities
A consortium called the Sequencing Alliance for Food Environments (SAFE) was launched on April 26, 2016, at University College Dublin (UCD) as a partnership of Dairygold, Cork, Ireland (dairygold.ie); Dawn Farm Foods, Kildare, Ireland (dawnfarms.ie); Glanbia, Kilkenny, Ireland (glabia.com); Kerry Group, Tralee, Ireland (kerrygroup.com); Mead Johnson Nutrition, Glenview, Ill. (meadjohnson.com); Nutrition Supplies, Cork, Ireland (nutritionsupplies.ie); and Creme Global, Dublin, Ireland (cremeglobal.com). Researchers at UCD will track the environments in a number of the partners’ food manufacturing plants in Ireland over a two-year period, mapping the microbiomes of the facilities. The consortium will use gene sequencing technology and statistical analysis to define bacterial characteristics at the DNA level and develop predictive software that will enable faster and more accurate quality control of the bacteria present in food facilities.
The Institute for Food Safety and Health (IFSH) at the Illinois Institute of Technology, Chicago initiated a WGS project in early 2015 with representatives from several food companies. Behzad Imanian, project leader since March 2016, said that the project intends to promote and implement WGS technology in identifying, differentiating, and tracing foodborne organisms back to their original sources. It has become the most reliable and fastest technology, he said, and is more economical than the older technologies used by food companies. The project promotes collaboration among members of the food industry and regulatory agencies. In the project, food companies submit samples containing known or unknown microorganisms, and the IFSH sequencing and bioinformatics teams process the samples, obtain sequence data, assemble and analyze the sequences, and provide data about the samples to the food companies and to the GenomeTrakr database.
The Grocery Manufacturers Association (GMA) has been monitoring the interest in WGS among its food company members. Melinda Hayman, GMA’s director of microbiology, said that changes in food production and supply such as centralizing processes, importing foods year round, and globalization intensify food safety issues. Detecting pathogens quickly and accurately is paramount, and in deciphering genetic information down to each individual nucleotide in the DNA, WGS provides higher reliability of an organism’s identity by discriminating below the species level. She said that the GMA’s microbiological safety committee plans to create a white paper on the practical applications of WGS in the near future.
Sequencing Instrumentation
Companies offering instrumentation for conducting whole genome sequencing include the following:
• Illumina, San Diego, Calif. (illumina.com), offers MiniSeq, MiSeq, NextSeq 500, and HiSeq 2500, 3000, and 4000 sequencers that are said to provide unparalleled raw read accuracy, read length, and read depth for high-quality draft and complete microbial genome assemblies.
• 454 Life Sciences, Basel, Switzerland (454.com), offers the Genome Sequencer FLX and GS Junior systems for high-throughput DNA sequencing, providing long, highly accurate sequence reads, including paired-end reads.
• Pacific Biosciences, Menlo Park, Calif. (pacb.com), offers PacBio Sequencing Systems that use single-molecule, real-time sequencing technology to provide a comprehensive view of genomes, transcriptomes, and epigenomes.
• Thermo Fisher Scientific, Waltham, Mass. (thermofisher.com), offers Ion Torrent, Ion S5, Ion S5 XL, and Ion Proton systems as well as the Ion PGM system for whole genome sequencing of microbes.
Correction
In the May 2016 issue of Food Technology, the food safety and quality column, “Calories Count on Menus,” incorrectly indicated that a Eurofins facility in Horsham, Pa., provides chemical analysis for nutrition labeling. The Eurofins Nutrition Analysis Center, Des Moines, Iowa (eurofins.com), provides that service.
 Neil H. Mermelstein, IFT Fellow,
Neil H. Mermelstein, IFT Fellow,
Editor Emeritus of Food Technology
[email protected]


