Microbial Modeling Predicts Shelf Life and Safety
FOOD SAFETY AND QUALITY
 Microbial modeling is not a microorganism walking down a runway like a fashion model but rather a method of predicting the shelf life and safety of food formulation options. It is being increasingly used by food companies and regulatory agencies to develop products and processes, set performance standards, evaluate regulatory compliance, and assess risk. In microbial modeling, mathematical models are used to predict the growth or survival of microorganisms in foods. The models are developed by studying the growth or survival of microorganisms over a period under specified conditions of storage. The data are then statistically fitted to equations that can be used to predict the microbial growth or survival under other conditions. It is a way for researchers, food processors, and regulatory agencies to predict the effect of potential changes to formulations or environmental conditions on microbial growth or survival during production, transportation, and storage. It allows food industry professionals to narrow potential formulations and conduct final studies with the best formulation. Proper data collection and analysis are key to successful application of predictive modeling.
Microbial modeling is not a microorganism walking down a runway like a fashion model but rather a method of predicting the shelf life and safety of food formulation options. It is being increasingly used by food companies and regulatory agencies to develop products and processes, set performance standards, evaluate regulatory compliance, and assess risk. In microbial modeling, mathematical models are used to predict the growth or survival of microorganisms in foods. The models are developed by studying the growth or survival of microorganisms over a period under specified conditions of storage. The data are then statistically fitted to equations that can be used to predict the microbial growth or survival under other conditions. It is a way for researchers, food processors, and regulatory agencies to predict the effect of potential changes to formulations or environmental conditions on microbial growth or survival during production, transportation, and storage. It allows food industry professionals to narrow potential formulations and conduct final studies with the best formulation. Proper data collection and analysis are key to successful application of predictive modeling.
Symposium Addresses Modeling
Last year at IFT17, speakers in the symposium “Development and Applications of Microbiological Models” discussed the principles of predictive microbiology and demonstrated how to develop and use predictive models. Cheng-An Hwang, research food technologist, Residue Chemistry and Predictive Microbiology Research Unit of the U.S. Dept. of Agriculture’s Agricultural Research Service (USDA-ARS), discussed the development of microbial models and their applications in food manufacturing and microbial risk assessment. In predictive microbiology, he said, microbial growth, survival, and inactivation can be quantified and expressed with mathematical equations and can be reproduced under a specific set of environmental conditions. He described the step-by-step procedure for developing microbial models.
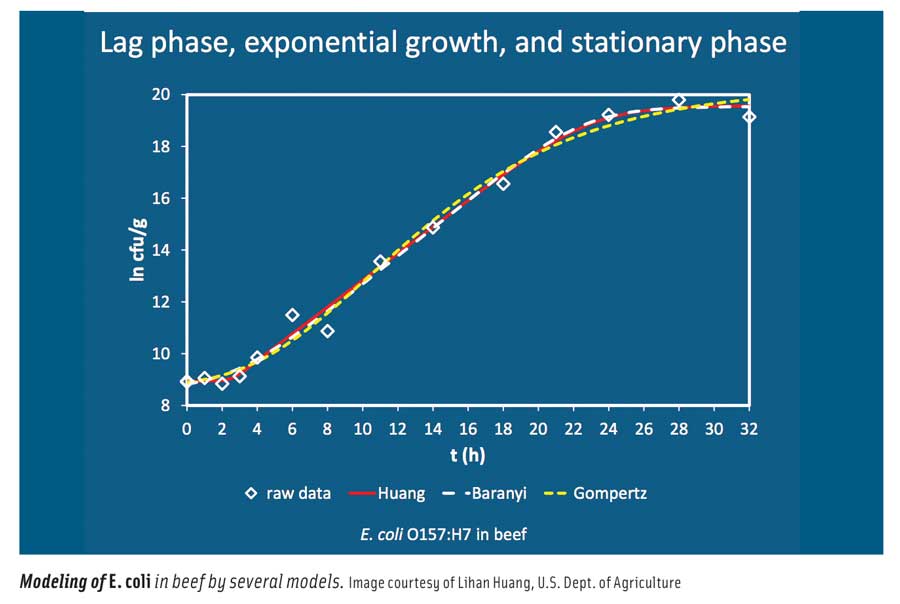
In general, he said, mathematical models used in predictive microbiology can be categorized into primary, secondary, and tertiary models. Primary models describe microbial responses such as population growth and toxin development with time; secondary models describe the responses to changes in environmental conditions such as temperature, pH, and water activity; and tertiary models turn the primary and secondary models into application software. He described the various primary models (Gompertz, Baranyi, Buchanan, and Huang); secondary models (polynomial, square root, gamma concept, and cardinal parameters); and tertiary models, which are usually Excel spreadsheets, computer programs, or online applications. He illustrated his presentation with examples of Listeria monocytogenes in seafood salad; Escherichia coli O157:H7 in ground beef; and E. coli O157:H7, Salmonella, and L. monocytogenes in fermented dry and semi-dry sausage. He concluded by saying that microbial modeling can be used to identify potential microorganisms of concern, identify controls and levels, optimize formulations and processes, address regulatory performance standards, design HACCP plans, predict microbiological quality and safety, and assist in microbial risk assessment.
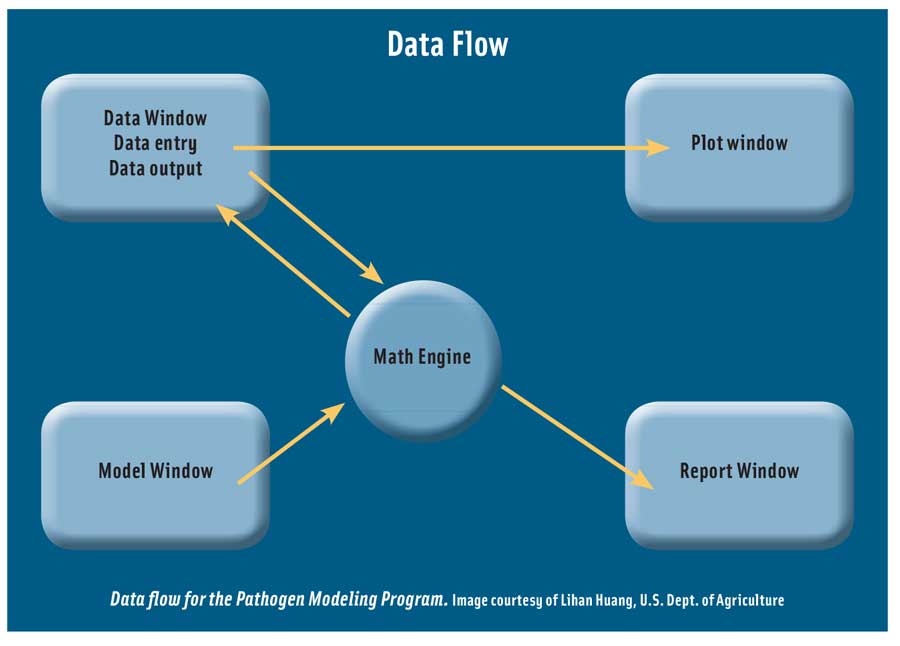
Lihan Huang, research leader, Residue Chemistry and Predictive Microbiology Research Unit, USDA-ARS, described the Integrated Pathogen Modeling Program (IPMP) he developed. He said that mathematical modeling and data analysis used to be a very challenging task that required relatively advanced knowledge of mathematics, computer programming, and statistics. He developed IPMP to meet the needs of food scientists and microbiologists who want to learn and apply predictive microbiology but feel inadequate in mathematics and programming. The IPMP is a user-friendly tool that allows anyone who is interested in predictive modeling to comfortably develop predictive models, he said. It is a suite of data analysis tools that contains more than 20 of the most frequently used primary and secondary models. It features easy-to-use graphical-user interfaces to guide every step, along with standardized data analysis and interpretation.
Huang described the various models used to analyze microbial growth; data analysis techniques, including linear regression and nonlinear regression; and typical problems. He said that commercial data analysis and statistical tools are very powerful but require product-specific programming, require advanced training, are difficult to learn and use, and are expensive. Potential users may feel that the math is too hard, don’t know which model to choose or which model is better, don’t have a program to do predictive modeling, or don’t know how to program. The IPMP overcomes these problems, he said. He illustrated its use with data on Cronobacter sakazakii in reconstituted infant formula and L. monocytogenes in beef frankfurters and ground beef. The parameters estimated by IPMP, he said, are identical or equivalent to those of SAS and R programs. The IPMP, he summarized, has significantly lowered the bar for those who want to work on predictive modeling, he said, and the food industry can use this tool to develop models for microbial shelf-life prediction.
Emma Hartnett, director, risk assessment of biological systems, Risk Sciences International (risksciences.com), described the FDA-iRISK online quantitative risk assessment system that allows users to estimate and compare the risk of foodborne illness from microbial and chemical hazards and the public health impact of interventions. It enables users to build, view, and share scenarios that reflect their actual or theoretical food safety issues without requiring extensive risk-assessment modeling experience.
The user creates a scenario by entering data for elements such as food, hazard, population, process model, consumption model, dose response, number of cases, and population burden into a built-in model framework. The system integrates the data using built-in mathematical calculations and predicts public-health outcomes. The user can make changes to the scenario data to ask, “What if?” She illustrated use of the system with apple juice data from a draft U.S. Food and Drug Administration (FDA) risk assessment and with a hypothetical event during handling and storage in the home, namely, rare cross-contamination of L. monocytogenes from a cutting surface to cantaloupe.
Public-health outcomes can be compared by such values as disability-adjusted life years (DALY), quality-adjusted life years, and cost of illness. For example, she said, imagine hazard A causing two fatalities and hazard B causing 100,000 cases of gastroenteritis with 10% long-term disability. Which incurs the larger burden of disease? How does one compare morbidity with mortality? She said that fatal and nonfatal outcomes can be compared using the DALY, for example. One DALY is equivalent to one person dying a year short of his life expectancy, two people dying six months early, five people suffering a 20% loss of function lasting one year, or one person dying six months early and another suffering a 50% loss of function lasting one year.
Modeling Tools Available
Predictive Microbiology Information Portal, developed by the USDA-ARS and the USDA’s Food Safety and Inspection Service, links users to the most comprehensive websites associated with models, databases, regulatory requirements, and food safety principles. Intended to help food companies access information on the use of predictive models, their appropriate application, and proper model interpretation, it provides an overview of predictive modeling, tutorials, and a resource locator for information on 154 organisms and 155 food types.
Pathogen Modeling Program, developed by the USDA-ARS Residue Chemistry and Predictive Microbiology Research Unit, is a package of tertiary models that can be used to predict the growth and inactivation of bacteria, primarily foodborne pathogens, under various environmental conditions. Probably the most widely used predictive microbiology application software, it is regularly updated and expanded. The present version includes more than 40 models for different bacterial pathogens. The software allows growth or inactivation of pathogens to be predicted for different combinations of constant temperature, pH, level of salt, water activity, and, in some cases, other conditions such as organic acid type and concentration, atmosphere, or nitrate. It also includes models that predict the effect of cooling temperature profiles on growth of Clostridium botulinum and Clostridium perfringens after cooking.
IPMP is an easy-to-use, data analysis tool that allows anyone who is interested in predictive modeling to comfortably develop predictive models (primary and secondary). IPMP-Dynamic Prediction is an extension of the IPMP. Whereas the IPMP is designed to analyze individual growth or inactivation curves, the IPMP-Dynamic Prediction is used to predict the growth and inactivation of microorganisms exposed to dynamically changing temperature and other environmental conditions. A typical application is predicting the growth of C. perfringens in cooked meats during cooling.
IPMP-Global Fit, another extension of the IPMP, is designed as a one-step, direct kinetic analysis tool that constructs a tertiary model from the entire experimental data set of isothermal microbial growth and inactivation. It uses (fits) the entire set of experimental data for both primary and secondary models and obtains the kinetic parameters that minimize the global error of the primary model. It differs from the IPMP in that it analyzes all isothermal growth or inactivation curves obtained under different conditions together and is designed to derive kinetic parameters that are optimized for the entire data set. It includes four primary models for growth and survival and six secondary models.
FDA-iRISK, developed by the FDA’s Center for Food Safety and Applied Nutrition, the Joint Institute for Food Safety and Applied Nutrition, and Risk Sciences International, is designed to analyze data on microbial and chemical hazards in food and provide an estimate of the resulting health burden on a population. The input data include the food and its consumption data and processing/preparation methods, the hazard and its dose-response curve, and the anticipated health effects of the hazard when ingested by humans. It enables users to conduct quantitative, probabilistic risk assessments and build scenarios that mathematically simulate food safety issues at any point in the food chain. Users can assess the impact of interventions by varying their data to explore how changes in various practices in the food chain would be likely to affect public-health outcomes. In developing a risk-ranking toolbox, the European Food Safety Authority has stated that FDA-iRISK was the most appropriate tool for risk ranking of microbiological hazards.
ComBase is a database developed by the USDA-ARS, the United Kingdom Institute of Food Research, and the Australian Food Safety Centre of Excellence that contains thousands of records for pathogenic and spoilage bacteria under many different environmental conditions. To search the database, users input criteria such as a specific microorganism, type of food, level of acidity, temperature, water activity, or salt concentration, and specific food conditions, such as additives, preparation methods, and packaging atmosphere. The database includes more than 50,000 curves of growth, survival, or inactivation of microorganisms in foods, including models for growth or inactivation of 18 foodborne pathogens and two spoilage bacteria (Brochothrix thermosphacta and Pseudomonas).
Food Spoilage and Safety Predictor is software developed by the National Food Institute, Technical University of Denmark, to facilitate the use of mathematical models to predict growth of spoilage and pathogenic microorganisms in food and the effect of constant or fluctuating temperature storage conditions on product shelf life. It includes models to predict the simultaneous growth of L. monocytogenes and lactic acid bacteria in chilled seafood, meat products, and cottage cheese and a generic model to predict growth of microorganisms in food depending on temperature; water activity or salt; pH; carbon dioxide; smoke intensity; nitrite; and acetic, benzoic, citric, lactic, and sorbic acids.
Forecast, a system offered by Campden BRI (campdenbri.co.uk), provides microbial modeling of spoilage organisms. A white paper on predictive microbiological models describes the system and includes models of spoilage organisms for fish, meat, fresh produce, and yeasts in fruits and drinks, plus models for acidified foods.
Risk Ranger is a food safety risk calculation spreadsheet from the Tasmania Institute of Agriculture’s Food Safety Centre that helps in determining relative risks from different product, pathogen, and processing combinations. It can be used by risk managers and others without extensive experience in risk modeling. The user is required to answer 11 questions related to the severity of the hazard, likelihood of a disease-causing dose of the hazard being present in a meal, and the probability of exposure to the hazard.
Sym’Previus, a microbiological data prediction tool, includes a database with growth and inactivation responses of microorganisms in foods and predictive models for growth and inactivation of more than 40 pathogenic and spoilage microorganisms.
 Neil H. Mermelstein, IFT Fellow, Editor Emeritus of Food Technology
Neil H. Mermelstein, IFT Fellow, Editor Emeritus of Food Technology
[email protected]