Moving Forward with Nutrigenomics
Providing valid personalized nutritional advice to consumers on the basis of their genetic makeup will require scientific collaboration, data sharing, and joint funding strategies.

Information obtained with genetic and emerging “omic” tests are personalizing medical treatments and drug selection (Kaput et al., 2007b), and the same was predicted for the advent of personalized nutritional advice (DeBusk et al., 2005; Ordovas, 2004), perhaps leading to the development of healthier processed foods targeted to individuals (Fogg-Johnson and Kaput, 2003).
In the early 2000s, companies rushed to the consumer market with genetic test offerings, reportedly linked to nutritional recommendations, to capitalize on “nutrigenomics.” The promise was genetic testing to enable personalized nutritional advice for consumers to avoid or mitigate chronic disease. In general, the test formats involved providing a sample of DNA, via cheek swab, and responding to questions about diet and lifestyle. The outputs of those tests fell short of the promise of “individualized” nutritional advice designed to prevent chronic diseases, including cardiovascular disease, osteoporosis, mental decline, and others.
Well-publicized hearings by the U.S. General Accountability Office (Kutz, 2006) reported on investigations of personalized genetic tests, testing companies, resulting nutritional advice, and cost of tests and recommended supplements. The investigation found that “results from all the tests GAO purchased misled consumers by making predictions that are medically unproven and so ambiguous that they do not provide meaningful information to the consumer.” The GAO report also noted that the nutritional advice provided was highly generalized and nonspecific. Several nutrigenetic testing companies recommended expensive supplements that were more costly than similar products generally available to the public.
With the ever-evolving inventory of peer-reviewed scientific studies showing associations between nutrients, genetic makeup, and health outcomes, one has to wonder why the promise was not delivered. Indeed, the question now is whether personalized nutrition can be delivered meaningfully and profitably.
While many, including us, believe the goals of personalized nutritional advice based on genetic makeup will be realized, expectations have given way to the sobering recognition of the challenges of understanding genetic causes of complex traits such as chronic diseases and the role of environment, including diet, in regulating expression of genetic makeup. This understanding has lead to refinement in thinking and approaches to implementation of tools to deliver the promise of personalized nutrition.
An Emerging Discipline
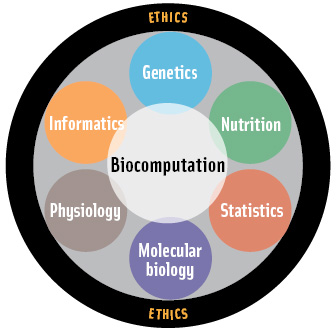
While both nutritional and genetic sciences continue to make fundamental contributions to disease prevention and treatments, a more complete understanding of health and disease processes must include simultaneous analyses of nutrient intakes and genetic makeups. Nutrigenomics differs conceptually and practically from nutritional and genetic research and applications in that both genotype and environment are assessed in each nutrigenomics experiment. Such studies have become feasible only since the advent of genomic (genetic and transcriptomic), proteomic, and metabolomic technologies that can analyze thousands of analytes simultaneously.
--- PAGE BREAK ---
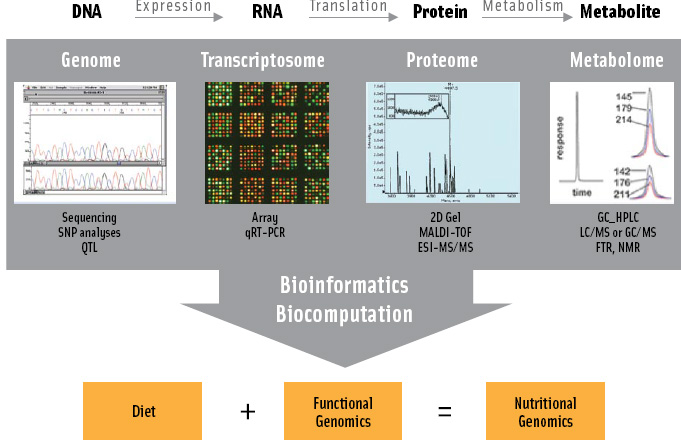 As in any emerging field, new and confusing definitions abound. For simplicity, nutrigenomics is best considered a conceptual approach to scientific research that analyzes genetic makeup and nutrient intakes with the understanding that other environmental factors (e.g., physical activity, exposure to toxins) cannot be ignored and should be measured if possible. The technologies employed in nutritional genomic research are identical to functional genomics—high-throughput analyses of genotype (gene variants, ancestral background), transcripts, proteins, and metabolites, i.e., “omics technologies.” Bioinformatics, defined as acquisition, management, storage, and retrieval of high-throughput datasets, and biocomputation, defined as the analyses of those datasets, are critical components of the research and application of nutrigenomic knowledge.
As in any emerging field, new and confusing definitions abound. For simplicity, nutrigenomics is best considered a conceptual approach to scientific research that analyzes genetic makeup and nutrient intakes with the understanding that other environmental factors (e.g., physical activity, exposure to toxins) cannot be ignored and should be measured if possible. The technologies employed in nutritional genomic research are identical to functional genomics—high-throughput analyses of genotype (gene variants, ancestral background), transcripts, proteins, and metabolites, i.e., “omics technologies.” Bioinformatics, defined as acquisition, management, storage, and retrieval of high-throughput datasets, and biocomputation, defined as the analyses of those datasets, are critical components of the research and application of nutrigenomic knowledge.
While nutrigenomics encompasses the full spectrum of research strategies from basic cellular and molecular biology to whole-body metabolism, clinical science, and population health, the key aspects of the experimental designs are the analyses of genotypes and nutrient intakes in humans or the systematic variation of genotype and nutrient intakes in model systems such as cell culture and laboratory animals.
A large number of studies and resulting publications have documented that nutrients alter the expression of genetic information at the level of gene regulation and signal transduction, and through alterations of chromatin structure, enzymes, and proteins (reviewed in Kaput and Rodriguez, 2004). In addition, nutrigenetic population studies have shown reproducible gene–nutrient–phenotype associations demonstrating that individuals with different alleles in genes are affected differently by nutrient intakes (reviewed in Corella and Ordovas, 2005). Hence, the scientific concepts acknowledged since Hippocrates uttered “let food be your medicine and medicine your food” have now been proven with modern technologies at the molecular genetic level.
The significant progress and excitement of these early studies is tempered because gaps continue to emerge in our understanding of health and disease processes in relation to diet. These gaps include how multiple genes and many nutrients interact in each individual and the best approaches for applying nutrigenomic concepts, results, and knowledge for personalized nutrition and medicine and for improving public health. Hence, there is a continued need for basic scientific research integrating nutrition, genetic, physiology, public health, and related disciplines.
Pharmacogenomics, toxicogenomics, and nutrigenomics research and applications must contend with the same gaps: (1) genetic heterogeneity of humans, (2) complexity of foods and the interactions of dietary chemicals with molecular processes, and (3) intricacies of metabolic pathways and networks underlying health and disease, i.e., the diversity challenges (Kaput et al., 2007a).
Results from genetic studies attempting to associate a gene or its variants to a disease or specific metabolic phenotype illustrate the challenges: many published gene–disease associations could not be replicated in subsequent studies. This led one accomplished epidemiologist to publish a report entitled “Why most published research findings are false” (Ioannidis, 2005), identifying experimental designs where sample sizes lacked appropriate statistical power, control groups were not appropriately matched to cases, study participants of differing genetic admixtures (populations stratification), and data that were overinterpreted (e.g., Newton-Cheh and Hirschhorn, 2005). Researchers working at the intersection of nutrition and genetics added that diet–gene interactions are major contributors to the control of gene expression and could influence associations among genes, phenotype, and dietary intake (Ordovas and Corella, 2006).
Reaching the goal of personalized nutrition will require an understanding of the scientific challenges of identifying nutrient–gene interactions involved in health or disease development and realistic solutions to surmount those challenges.
--- PAGE BREAK ---
Scientific Diversity Challenges
Humans have adapted to almost every nutritional environment on earth, from northern tundra to equatorial, and from deserts to high-altitude ecosystems. One could conclude that as long as minimum amounts of energy and micronutrients were consumed from the varying flora and fauna in these different regions, the most significant contribution to health and disease susceptibility would be one’s genetic makeup. The apparent adaptability, however, masked the effects of nutrients on the expression of genetic makeup and the consequences of those interactions on the promotion of health or the development of chronic diseases (Kaput and Rodriguez, 2004).
The most obvious example of the importance of environment on disease is the rise in the incidence of obesity and its complications (www.cdc.gov/nccdphp/dnpa/obesity/) during the past 20 years. Genetic changes to such large numbers of individuals could not have occurred so rapidly to cause such rapid weight gains on a population level. Rather, changes in diet and physical activity habits have produced the epidemic of obesity, type 2 diabetes, and their complications. This conclusion is supported by traditional epidemiological and immigration studies (e.g., (Kolonel et al., 2004)) that associate environment with chronic diseases. However, the underlying molecular mechanisms cannot be ascertained by statistical associations derived from population studies, since such research links the average nutrient intakes with the average physiological response of everyone in the study.
 Numerous in-vitro, animal, and human studies have demonstrated that chemicals classified as macronutrients (certain fatty acids) and bioactives such as genistein, sterols, and micronutrients regulate gene expression directly (reviewed in Schuster, 2006) or through changes in signal-transduction pathways (Guo and Sonenshein, 2006). Hence, for the specific chronic disease of obesity, overconsumption of calories (Goldberg et al., 2004) and subsets or specific bioactive chemicals (e.g., Jequier, 2002) alter the regulation of genes that cause increased weight deposition in genetically susceptible individuals.
Numerous in-vitro, animal, and human studies have demonstrated that chemicals classified as macronutrients (certain fatty acids) and bioactives such as genistein, sterols, and micronutrients regulate gene expression directly (reviewed in Schuster, 2006) or through changes in signal-transduction pathways (Guo and Sonenshein, 2006). Hence, for the specific chronic disease of obesity, overconsumption of calories (Goldberg et al., 2004) and subsets or specific bioactive chemicals (e.g., Jequier, 2002) alter the regulation of genes that cause increased weight deposition in genetically susceptible individuals.
In addition to chemicals present in unprocessed foods, our diets are now greatly influenced by modern food production methods. The U.S. Dept. of Agriculture reported that about 75% of all retail foods in the world were processed foods (www.ers.usda.gov/publications/aib794/aib794.pdf). While many of these manufactured foods may be nutritionally balanced, the amount of salt, processed carbohydrates, and unbalanced fat (i.e., unbalanced omega-3 to omega-6 fatty acid ratio) may be contributing to the worldwide epidemic in obesity and its complications (e.g., Brennan, 2005).
Regardless of the type of food, identifying the specific pathways and the effect of different amounts of these bioactive chemicals is a critical part of understanding how diet maintains health or leads to disease in each individual. For more complete analyses of gene–disease (or gene-phenotype) associations, studies must assess food intake as well as genetic differences. Accurately assessing nutrient intake in epidemiological studies is challenging, since free-living humans do not regard daily life as a science experiment where the amount and type of food are accurately and precisely measured and recorded. The nutrition community generally acknowledges that food-frequency questionnaires (FFQs) for assessing intakes are less than ideal (Rutishauser, 2005). Nevertheless, FFQs are likely to be the most convenient assessment for large-scale studies. Quantitative biological measurement tools for nutrient intakes need to be developed for more accurately assessing nutrient intake (Corella and Ordovas, 2005).
A related challenge is converting food intake to nutrients, usually done using food composition databases. The most complete is the USDA food composition database (www.nal.usda.gov/fnic/foodcomp/Data/SR18/sr18.html), although the European Union has developed and continues to improve the comparable European Food Information Resource Network EuroFir database (www.eurofir.net). However, plant species, like other organisms, respond to their environments, which may result in different nutrient contents depending on the genotype of the plant and its growth conditions (Reynolds et al., 2005). Since a full nutrient profile costs about $2,000 U.S. per sample in 2002 (Haytowitz et al., 2002) and it is typical to average six samples, a ranking scheme will be needed to prioritize analyses of foods.
--- PAGE BREAK ---
Progress in developing national and international food composition databases is being made, as noted by the International Life Sciences Institute (ILSI) Crop Composition Database www.cropcomposition.org/), the International Network of Food Data Systems (www.fao.org/infoods/directory_en.stm), and the aforementioned EuroFir database.
An added layer of complexity occurs when comparing across populations. Cultural differences in food manufacturing, preparation, and eating customs exist not only among nations and ethnic groups but also among religions (e.g., Kim and Sobal, 2004). Personal choices such as various vegetarian practices, sleep time and continuity, activity levels, and other factors (references in Kaput et al., 2005) will also confound assessments of nutrient intakes. Individual studies have successfully included one or more of these variables, but to date, no study has been published with complete environmental analyses.
Genetic Diversity Challenges
Africans have the greatest variation in DNA sequence compared to other populations, a result consistent with the hypothesis that modern humans evolved on that continent (Jorde and Wooding, 2004). Groups of individuals carried subsets of the genetic diversity within a population during migrations from east Africa and subsequent population centers to each new location. Food availability and other factors contributed pressures for selecting and maintaining specific gene variants in new environments.
Different alleles conferring the same phenotype (e.g., lactase persistence) have practical consequences for both pharmacogenomics and nutritional genomics: gene variant–phenotype associations may be population specific because members of different ancestral groups may have different polymorphisms affecting the same gene; for example, the European allele of the lactase gene encodes a thymidine (instead of cytosine) at -13910 from the start of the lactase gene, but in certain African populations, the relevant polymorphism (cytosine instead of guanine) occurs at position -14010 from the start of the lactase reading frame (Tishkoff et al., 2007).
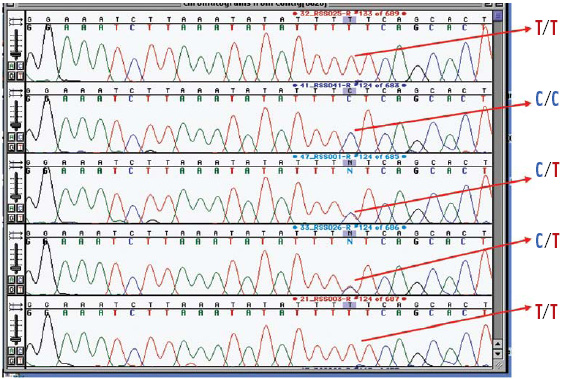 The implications of this genetic similarity and diversity are also significant for nutrigenomic researchers, since the frequency of gene variants differs among populations. Perhaps as important, gene variants and their products (either RNA or DNA) interact with other genes and their products in pathways and networks. Such gene–gene interactions, or epistasis (Carlborg and Haley, 2004), work at the protein and enzyme—and probably iRNA (Nakahara and Carthew, 2004)—levels, which sometimes counteracts deleterious alleles. A related concept is biochemical buffering—the ability of a system of biochemical reactions to produce similar fluxes through pathways to maintain homeostasis (Hartman, 2006). The concept of buffering is important because the presence or absence of a particular gene variant (i.e., single nucleotide polymorphism, SNP) is not an all-or-nothing phenomenon. The result of epistasis and biochemical buffering is that a single SNP is not deterministic but expresses itself within the background of the individual’s genome (i.e., the total variations in their DNA). Hence, SNP analyses of individual genes must be accompanied by analyses of all “relevant” genomic variations, an analytical approach which has yet to be developed.
The implications of this genetic similarity and diversity are also significant for nutrigenomic researchers, since the frequency of gene variants differs among populations. Perhaps as important, gene variants and their products (either RNA or DNA) interact with other genes and their products in pathways and networks. Such gene–gene interactions, or epistasis (Carlborg and Haley, 2004), work at the protein and enzyme—and probably iRNA (Nakahara and Carthew, 2004)—levels, which sometimes counteracts deleterious alleles. A related concept is biochemical buffering—the ability of a system of biochemical reactions to produce similar fluxes through pathways to maintain homeostasis (Hartman, 2006). The concept of buffering is important because the presence or absence of a particular gene variant (i.e., single nucleotide polymorphism, SNP) is not an all-or-nothing phenomenon. The result of epistasis and biochemical buffering is that a single SNP is not deterministic but expresses itself within the background of the individual’s genome (i.e., the total variations in their DNA). Hence, SNP analyses of individual genes must be accompanied by analyses of all “relevant” genomic variations, an analytical approach which has yet to be developed.
Epistasis was shown for a specific group of SNPs (called a haplotype, in this case HapK) in leukotriene A4 hydrolase (LTA4H), which was associated with increased risk of myocardial infarction (MI). African-Americans who carried the HapK haplotype derived from Europe (Helgadottir et al., 2006) had a much greater risk of MI and cardiovascular events than Europeans who carried HapK. The variants of one or more genes present in Africans affected the activity of the European-derived LTA4H variant more than European variants of the same genes. These results demonstrate the concept that a SNP or haplotype is “context specific” (Klos et al., 2005), i.e., that an SNP or haplotype may contribute different amounts to disease risk, depending on ancestral background because of epistatic (gene–gene) interactions.
Since diet is known to influence pathways involved in disease initiation or progression, the same nutrients may differentially affect individuals with different ancestral backgrounds, depending on the variants of the genes that are typical of the ancestral background. Note however, that while certain alleles may be found more frequently in one or another ancestral group, the variation within a population is greater than between populations (Shriver et al., 2005). There will likely be no “ancestral group” diet.
--- PAGE BREAK ---
Complexity of Datasets
While these challenges seem intractable, high-throughput technologies of genomics, transcriptomics, proteomics, and metabolomics will produce large datasets (also called high dimensional data) of nutrient intakes, gene variants, genetic ancestry data, physiological measurements of proteins and metabolites, as well as physical activity levels and other personal habits. Patterns among these datasets will then have to be extracted to produce knowledge of gene–environment interactions for each individual. The existing paradigm of analyzing biological data often assumes linear relationships, such as dose-dependent effects. However, genes, nutrients, and their interactions may not necessarily be related linearly (Calabrese and Baldwin, 2003).
Other disciplines have developed algorithms that can discover nonlinear patterns in highly dimensional data. Dimensionality reduction is a mathematical method of mapping multidimensional data into a space of fewer dimensions, generating a deeper understanding of complex biological processes (e.g., Dawson et al., 2005). Among these methods are isomap (Roweis and Saul, 2000; Tenenbaum et al., 2000), factor analysis (www.chass.ncsu.edu/garson/pa765/factor.htm) and classification tree analysis (www.statsoft.com/textbook/stclatre.html), all of which can be used to analyze complex patterns of gene variants and their influence on subphenotypes (insulin levels or fasting glucose levels, etc.) in response to different dietary intakes (Ritchie, 2005). The behavior of interacting genetic and environmental systems cannot yet be predicted accurately, hence multiple approaches may be needed to extract knowledge from experimental data. Nevertheless, data generated from different high-throughput technologies may be analyzed with more confidence when other paradigms (e.g., nonlinearity) are considered possible.
Hence, the new paradigm emerging in the 21st century for biological research is based on the model of the large-scale genome projects, with the inclusion of nutritionists, food scientists, sociologists, and public health experts.
Harnessing Nutrigenomics for Health
Many scientists doing research at the turn of the millennium were taught to reduce the complexity of an experiment by examining small components of a larger system and to vary only one component at a time. This “reductionist” philosophy and its associated methods have contributed immensely to scientific progress and will always be needed to understand individual pathways and reactions of complex networks. However, reductionism cannot fully explain complex, interacting systems, since expression of genetic information relies on environmental factors and both components and their interactions change during development and aging. Such complexity is rarely accounted for in reductionistic experimental designs.
Nutrigenomics projects, therefore, will require best practices developed by experts in different disciplines. The European Nutrigenomics Organization (NuGO, www.nugo.org) began this process in early 2004 and has generated best-practices papers in transcriptomics (Garosi et al., 2005), metabolomics (Gibney et al., 2005), and proteomics (Fuchs et al., 2005), and ethical practices for conducting human studies (Bergmann et al., 2006). Additional needs are to develop more accurate quantitative food measurements and better biomarkers of food exposure in the healthy and disease states. Perhaps the biggest challenge will be a means to apply population-based risk factors for nutrient exposures to each individual, thereby realizing the promise of personalized nutrition. It is not likely that nutrigenomics will fulfill its promise of personalized nutrition without comparing responses of gene–nutrient interactions across many different genetic makeups.
Understanding gene–nutrient interactions and applying that knowledge for personal and public health will require strategic collaborations among academia, government, and industries, resource and data sharing, and joint funding strategies that cross discipline, organizational, commercial, and national boundaries (Kaput et al., 2005). Efforts to organize stakeholders in the international nutrigenomic community are underway with the expansion of NuGO into a Nutrigenomics Society. Developing personalized nutrition and thereby improving personal and public health will require such coordinated efforts.
Nancy Fogg-Johnson, a Professional Member of IFT, is Principal, Nutri+Food Business Consultants, 796 Panorama Rd., Villanova, PA 19085 ([email protected]). Jim Kaput is President, NutraGenomics. 7628 Garden Ln., Justice, IL 60458. Send reprint requests to author Fogg-Johnson. The preparation of this manuscript was supported by Grant MD 00222 from the National Center for Minority Health and Health Disparities Center of Excellence in Nutritional Genomics and EU FP6 NoE Grant, Contract No. CT2004-505944, from the European Union.References
Bergmann, M.M., Bodzioch, M., Bonet, M.L., et al. 2006. Bioethics in human nutrigenomics research: European Nutrigenomics Organisation workshop report. Brit. J. Nutr. 95: 1024-1027.
Brennan, C.S. 2005. Dietary fibre, glycaemic response, and diabetes. Molec. Nutr. Food Res. 49(6): 560-570.
Calabrese, E.J. and Baldwin, L.A. 2003. Hormesis: The dose-response revolution. Ann. Rev. Pharmacol. Toxicol. 43: 175-197.
Cardon, L.R. and Bell, J.I. 2005. Association study designs for complex diseases. Nat. Rev. Genet. 2001. 2(2): 91-99.
Carlborg, O. and Haley, C.S. 2004. Epistasis: Too often neglected in complex trait studies? Nat. Rev. Genet. 5: 618-625.
Corella, D. and Ordovas, J.M. 2005. Single nucleotide polymorphisms that influence lipid metabolism: Interaction with dietary factors. Ann. Rev. Nutr. 25: 341-390.
Dawson, K., Rodriguez, R.L., and Malyj, W. 2005. Sample phenotype clusters in high-density oligonucleotide microarray data sets are revealed using Isomap, a nonlinear algorithm. BMC Bioinformatics 6: 195.
DeBusk, R.M., Fogarty, C.P., Ordovas, J.M., et al. 2005. Nutritional genomics in practice: Where do we begin? J. Am. Dietet. Assn. 105: 589-598.
Englyst, K.N. and H.N. Englyst, H.N. 2005. Carbohydrate bioavailability. Brit. J. Nutr. 94(1): 1-11.
Ferguson, L.R. and Kaput, J. 2004: Nutrigenomics and the New Zealand food industry. Food New Zealand, 19: 29–36.
Fogg-Johnson, N. and Kaput, J. 2003. Nutrigenomics: An emerging scientific discipline. Food Technol. 57(4): 61–67.
Fuchs, D., Winkelmann, I., Johnson, I.T., et al. 2005. Proteomics in nutrition research: Principles, technologies and applications. Brit. J. Nutr. 94: 302-314.
Garosi, P., De Filippo, C., van Erk, M., et al. 2005. Defining best practice for microarray analyses in nutrigenomic studies. Brit. J. Nutr. 93: 425-432.
Gibney, M.J., Walsh, M., Brennan, L., et al. 2005. Metabolomics in human nutrition: Opportunities and challenges. Am. J. Clin. Nutr. 82: 497-503.
Goldberg, J.P., Belury, M.A., Elam, P., et al. 2004. The obesity crisis: Don’t blame it on the pyramid. J Am. Dietet. Assn. 104(7): 1141-1147.
Guo, S. and Sonenshein, G., 2006. Green tea polyphenols and cancer prevention. Chpt. 8 in Kaput and Rodriguez (2006), pp. 177–206.
Hartman, J.L. IV. 2006. Genetic and molecular buffering of phenotypes. In Kaput and Rodriguez (2006), pp. 105–134.
Haytowitz, D., Pehrsson, P.R., and Holden, J.M. 2002. The identification of key foods for food composition research. J. Food Composition Anal. 15: 183–194.
Helgadottir, A., Manolescu, A., Helgason, A., et al. 2006. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat. Genet. 38(1): 68-74.
Ioannidis, J.P. Why most published research findings are false. PLoS Med. 2(8): e124.
Jequier, E. 2002. Pathways to obesity. Intl. J. Obesity Related Metabol. Disorders 26(Suppl. 2): S12-S17.
Jorde, L.B. and S.P. Wooding, 2004. Genetic variation, classification and ‘race.’ Nat. Genet. 36(Suppl. 1): S28-S33.
Kaput, J. and Rodriguez, R.L. 2004. Nutritional genomics: The next frontier in the postgenomic era. Physiol. Genomics 16(2): 166-177.
Kaput, J. and Rodriguez, R.L. 2006. “Nutritional Genomics. Discovering the Path to Personalized Nutrition.” John Wiley and Sons, Hoboken, N.J.
Kaput, J., Noble, J., Hatipoglu, B., et al. 2005. The case for strategic international alliances to harness nutritional genomics for public and personal health. Brit. J. Nutr. 94(5): 623-632.
Kaput, J., Noble, J., Hatipoglu, B., et al. 2007a. Application of nutrigenomic concepts to type 2 diabetes mellitus. Nutr. Metab. Cardiovasc. Dis. 17(2): 89-103.
Kaput, J., Perlina, A., Hatipoglu, B., et al. 2007b. Nutrigenomics: Concepts and applications to pharmacogenomics and clinical medicine. Pharmacogenomics 8(4): 369-390.
Kim, K.H. and Sobal, J. 2004. Religion, social support, fat intake and physical activity. Public Health Nutr. 7(6): 773-781.
Klos, K.L.E., Kardia, S.L.R., Hixson, J.E., et al. 2005. Linkage analysis of plasma ApoE in three ethnic groups: Multiple genes with context-dependent effects. Annals Human Genet. 69: 157-167.
Kolonel, L.N., D. Altshuler, and B.E. Henderson, 2004. The multiethnic cohort study: exploring genes, lifestyle and cancer risk. Nat. Rev. Cancer 4(7): 519-27.
Kutz, G. 2006. Nutrigenetic testing: Tests purchased from four Web sites mislead consumers. U.S. General Accountability Office, Washington, D.C..
Nakahara, K. and Carthew, R.W. 2004. Expanding roles for miRNAs and siRNAs in cell regulation. Curr. Opinion Cell Biol. 16(2): 127-133.
Newton-Cheh, C. and Hirschhorn, J.N. 2005. Genetic association studies of complex traits: Design and analysis issues. Mutat. Res. 573(1-2): 54-69.
Ordovas, J.M. 2004. The quest for cardiovascular health in the genomic era: Nutrigenetics and plasma lipoproteins. Proc. Nutr. Soc. 63(1): 145-152.
Ordovas, J.M. and Corella, D. 2006. Gene-environment interactions: Defining the playfield. In Kaput and Rodriguez (2006), pp. 57–76.
Reynolds, T.L., Nemeth, M.A., Glenn, K.C., et al. 2005. Natural variability of metabolites in maize grain: Differences due to genetic background. J. Agric. Food Chem. 53: 10061-10067.
Ritchie, M.D. 2005. Bioinformatics approaches for detecting gene-gene and gene-environment interactions in studies of human disease. Neurosurg. Focus, 19(4): E2.
Roweis, S.T. and Saul, L.K. 2000. Nonlinear dimensionality reduction by locally linear embedding. Science 290: 2323-2326.
Rutishauser, I.H. 2005. Dietary intake measurements. Public Health Nutr. 8(7A): 1100-1107.
Schuster, G.U. 2006. Nutrients and gene expression. In Kaput and Rodriguez (2006), pp. 153–176.
Shriver, M.D., Mei, R., Parra, E.J., et al. 2005. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Human Genomics 2(2): 81-89.
Tenenbaum, J.B., de Silva, V., and Langford, J.C. 2000. A global geometric framework for nonlinear dimensionality reduction. Science, 290: 2319-2323.
International HapMap Consortium. 2005. A haplotype map of the human genome. Nature 437: 1299-1320.
Tishkoff, S.A., Reed, F.A., Ranciaro, A., et al. 2007. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39(1): 31-40.
