Testing for Salmonella
LABORATORY
A foodborne illness outbreak that began in April 2008 and spread over 43 states, the District of Columbia, and Canada made more than 1,430 people sick and may have led to two deaths. An intensive epidemiological investigation beginning in May by the Centers for Disease Control and Prevention in collaboration with the Food and Drug Administration, the Indian Health Service, and state public health officials identified the cause of the outbreak as the Saintpaul serotype of Salmonella enterica and the source of the illness as jalapeño and serrano peppers imported from Mexico. The investigation initially identified tomatoes as the source, but FDA later lifted its warning on consuming them. Updates can be found on CDC’s Web site (www.cdc.gov/salmonella/saintpaul) and FDA’s Web site (www.fda.gov/oc/opacom/hottopics/tomatoes.html).

One of the problems involved in correctly identifying the real culprit, according to Ahmed E. Yousef ([email protected]), Professor of Food Microbiology at The Ohio State University, Columbus, is that there are about 2,500 serotypes of Salmonella and many subtypes in each, so identifying the serotype is not enough. Simply finding the Salmonella serotype in a patient and a food is not 100% proof that this particular food is the cause of the disease outbreak, he said. We have to identify the specific subtype and show that the bacterium in the food is the same as that isolated from the people experiencing the illness. Isolates of the Saintpaul serotype have genetic variations, he said, so to link the food to the illness, we need to match the genetic fingerprint of Salmonella Saintpaul from both sources.
Techniques for Identification
Conventional microbiological methods used to detect and identify Salmonella serotypes may require up to 5–6 days, so rapid and automated methods based on serological and molecular approaches have been developed.
A fingerprinting technique called pulsed-field gel electrophoresis (PFGE) is CDC’s method of choice for epidemiologic subtyping of pathogenic bacteria. It produces unique DNA patterns that serve as bacterial fingerprints. CDC developed standardized PFGE methods and in collaboration with the Association of Public Health Laboratories (APHL) created a national database called PulseNet so that scientists at public health laboratories throughout the country could rapidly compare the fingerprint patterns of bacteria isolated from ill persons to those in the database to identify specific bacteria and aid in epidemiological investigations.
Laboratories participating in PulseNet perform DNA fingerprinting by PFGE on disease-causing bacteria isolated from humans and from suspected food using standardized equipment and methods. DNA from isolated bacteria is fragmented, and the resulting fragmentation patterns are compared. Non-conventional electrophoretic equipment facilitates the migration of large DNA fragments through agarose gels by constantly changing the direction of the electrical field during electrophoresis.
Yousef said that even though PFGE is a very reliable test, slight variations in the protocol can produce different patterns. Some irregularities in fragmentation patterns can result even if everyone follows the protocol agreed on. The PFGE protocol is also time-consuming, having a turnaround time of about five days. For these reasons, Yousef and Luis Rodriguez-Saona ([email protected]), Assistant Professor in OSU’s Dept. of Food Science and Technology, have been developing a rapid, high-throughput technique for detecting and differentiating Salmonella serotypes and subtypes. The technique—Fourier transform infrared (FTIR) microspectroscopy—is said to be easier, more reliable, and much faster than the PFGE technique. It seems to be less affected by variations in protocol, Yousef said, so it may be better than PFGE for interlaboratory comparisons.
--- PAGE BREAK ---
In FTIR analysis, the absorption of infrared radiation by the bacteria produces a pattern of bands—a spectrum—that is unique to the strain, which might be a serotype or a subtype of S. enterica. Rodriguez described the FTIR procedure as follows: A cell suspension of the test bacterium is filtered through a hydrophobic-grid membrane filter (HGMF). The filter is then incubated on a suitable agar medium for up to 24 hr and dried before being examined by an IR microscope. A germanium-tipped probe attached to the microscope produces a wave that interacts with bacterial cells on the filter and produces a spectrum. The spectra collected are subjected to principal-component statistical analysis, and statistical models called soft independent modeling of class analogy (SIMCA) models are developed. Analysis of the filter-mounted cells takes less than 15 min.
The limiting factor, Rodriguez said, is growing enough of the bacteria on the hydrophobic grid to reach a specific biomass (104–105 cells) that would give a reproducible signal. This usually takes 12–18 hr. Also, the growth media will influence the spectral fingerprint, and use of selective media has improved the differentiation among different isolates of S. enterica serotypes. The HGMFs are dried before examination because water produces a very high signal in IR that would mask the signal from the bacteria.
Rodriguez said that analysis of the spectra has shown well-defined clusters for each Salmonella serotype and differentiation among serotypes, including Saintpaul. He added that multivariate analysis has shown that lipopolysaccharides in the bacterial cell outer membrane are responsible for the differentiation among isolates.
IR technology is growing at a fast pace, he said. Various companies offer benchtop equipment for the technique, including Varian (www.varianinc.com), Thermo Scientific (www.thermo.com), Bruker Optics (www.brukeroptics.com), Shimadzu (www.shimadzu.com), and Jasco (www.jascoinc.com). Smiths Detection (www.smithsdetection.com) makes a one-piece IR microscope that transforms a commercial light microscope into a versatile infrared microprobe for molecular analysis. Rodriguez added that portable handheld infrared spectrometers equipped with an attenuated total reflectance (ATR) diamond crystal (www.ahurascientific.com) are becoming available and could provide direct fingerprinting strategies for rapid and specific microbial analysis.
What’s Needed
Daniel Y.C. Fung ([email protected]), Professor of Food Science at Kansas State University, Manhattan, said that advances in rapid detection of pathogens are occurring, but we still need to have very good statistical sampling procedures, as well as very good enrichment procedures to let Salmonella grow so we can detect it. We also need very good antibodies to type the serotypes. Polyclonal antibodies trap more antigens, but monoclonal antibodies are more specific of an organism’s antigen. We need monoclonal antibodies specific for the Saintpaul serotype.
With rapid methods, if we get a presumptive negative, we’re done, he said—there’s no Salmonella present. However, if we get a presumptive positive, then we still need to go through the conventional culture method to confirm it. The gold standard is the conventional method described in FDA’s Bacterial Analytical Manual (www.cfsan.fda.gov/~ebam/bam-toc.html); the method for Salmonella can be found in chapter 5 (www.cfsan.fda.gov/~ebam/bam-5.html).
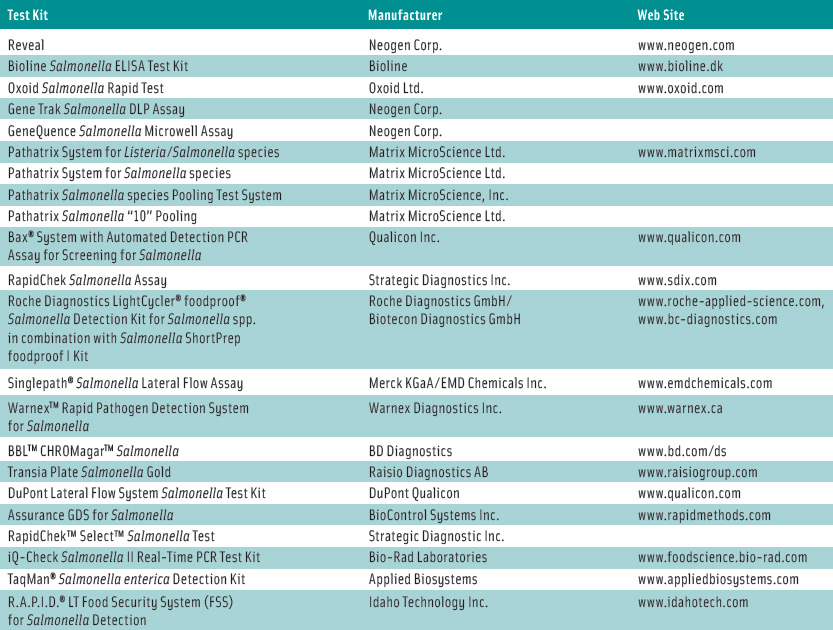
Rapid test kits are very reliable, he added. If a kit can detect the organism 90–95% of the time, it’s reliable. A number of test kits for Salmonella detection have received Performance Tested Methods certification from the AOAC Research Institute, which provides an independent third-party review of test kit performance claims. These methods are listed in Table 1.
--- PAGE BREAK ---
Fung briefly reviewed some of the approaches to detection and identification of microorganisms:
• Monoclonal and polyclonal antibodies are used to bind with antigens on the organism’s surface to detect and characterize microorganisms and their components. Some companies offer completely automated enzyme-linked immunosorbant assays (ELISAs). For example, the VIDAS system from bioMèrieux (www.biomerieux-usa.com) can automatically perform ELISAs in 45 min–2 hr after pre-enrichment of target organisms.
• Lateral-flow antigen–antibody tests, often referred to as dipstick tests, take only about 10 min after overnight enrichment.
• Immunomagnetic separation is being used to capture target molecules, reducing incubation time and increasing sensitivity. The Pathathrix system from Matrix Micro-Science Ltd. (www.matrixmsci.com), for example, recirculates a pre-enriched sample over antibody-coated paramagnetic beads for 30 min to capture as many target organisms as possible for subsequent detection by ELISA, PCR, culture, or other methods.
• Polymerase chain reaction (PCR) detects pathogens by amplifying the target DNA and detecting the target PCR products. The target products have conventionally been detected by gel electrophoresis, which is time consuming, but faster fluorescence techniques are now on the market (real-time PCR). For these rapid techniques, the more target DNA in the sample, the faster the results will be. For example, the Salmonella detection kit from Applied Biosystems (www.appliedbiosystems.com) uses real-time PCR to identify pathogens in food in less than 24 hr.
Fung has been conducting for the past 28 years Kansas State’s annual International Workshop/Symposium on Rapid Methods and Automation in Microbiology. At these eight-day workshops, attendees hear experts discussing the newest advances and participate in hands-on sessions with new equipment, techniques, and test kits without making commitments to purchase the equipment and systems. More than 4,000 scientists from 60 countries have participated in these workshops. The most recent workshop, held June 13–20, 2008, also featured a special day-long session showcasing presentations on cutting-edge developments in molecular biology. The next workshop will be held in Manhattan, Kan., on June 19–26, 2009.
by Neil H. Mermelstein,
a Fellow of IFT, is Editor Emeritus of Food Technology
[email protected]


