Fingerprinting Microorganisms
DNA techniques provide reliable, reproducible, and indisputable characterization of microorganisms used in food fermentations and other applications.
Fermentation is used throughout the food and beverage industries to produce bread, cheese, yogurt, wine, and beer, as well as a wide range of food ingredients, from citric acid to monosodium glutamate.
In these fermentations, it is crucial to have only the correct microbes, since contamination is extremely costly and potentially dangerous. In addition, since the microorganisms used for these processes are often valuable and proprietary to the companies that use them, it has become increasingly important to have the tools to identify them with great precision. Genetic fingerprinting of microorganisms does this.
Why DNA Fingerprinting Is Needed
There are several reasons why genetic typing techniques are needed:
• Safety and Quality. The food industry is increasingly investing heavily in safety and quality. Companies try to design and optimize their products/ processes to minimize the risk of contamination by unwanted organisms, sometimes pathogens and sometimes just minor mutants of the desired organism. Hazard analyses and quality control procedures can be designed more easily if it is possible to identify with precision which microbes are likely to create problems.
Genetic fingerprinting can be used to assess quality more accurately at the end of the production, after which a positive decision to release a product batch can be made. It can also be used to monitor the quality of starter batches in more detail and follow the fermentation process throughout. These improvements will help prevent unpleasant surprises at the end of the fermentation process. In addition, consistent quality in, for example, the ratio and type of organisms can provide a competitive edge in the functional food business.
Contamination with spoilage organisms ruins the product and costs manufacturers money. Sometimes, these organisms are well-known and occur commonly; hence companies know at once what actions to take to avoid ruining the fermentation. At other times, spoilage organisms are not easy to characterize, and the appropriate and immediate actions cannot be taken until considerable additional work has been done. A complete inventory of recurring spoilage organisms and plans for appropriate and rapid remedial responses will help reduce contaminated batches to a minimum.
• Process and Product Optimization. To remain competitive, manufacturers must continually keep abreast of the latest technological advances to improve their processes and ultimately their products. In the fermentation industry, this may mean selection and identification of more-efficient and more stable microorganisms that can function under a variety of conditions. Optimization of processes can reduce the probability of failures occurring either through the growth of opportunistic but useless mutants or through the inhibition of the growth and performance of the desired microorganism.
For example, in the brewing industry, existing techniques are often incapable of discriminating between closely related lager and beer yeast strains. DNA fingerprinting allows brewers to resolve the differences between their strains.
In terms of process, the characteristics of yeasts selected for brewing can vary considerably in bactericidal qualities, glucoamylase activity, SO2 formation, diacetyl production, ethanol tolerance, and flocculation characteristics. These factors ultimately affect the flavor, clarity, and many other quality characteristics of the final product. It is important that the genetic profiles of microorganisms or their mixes responsible for a successful product can be clearly and unambiguously identified.
In the same way, it is important to identify process conditions under which mutants are more likely to form. Current procedures to maintain the genetic consistency of strains often require the use of a new ampule of the stock strain after 5–6 generations, but whether this is optimal or merely an economic decision is not always clear or has not been investigated because of to the lack of appropriate technologies.
--- PAGE BREAK ---
• Regulatory Requirements and Intellectual Property Rights. Requirements by worldwide regulatory bodies for the definitive identification of the microorganisms in use in food/beverage systems are likely to become increasingly stringent as a wider variety of beneficial organisms are offered to consumers. For example, more-precise specification of the organisms used as probiotics in animal feed has already become required by government regulations.
When an organism has commercial value, its owner will want to name it as part of a process of recognizing its value, i.e., claim intellectual property rights such as by patenting it. This results in a trend toward more names for more organisms. For example, strains of lactobacilli used as starter cultures or probiotics have proliferated in the dairy industry. But names have little value if they are not especially well characterized to protect property rights. Organisms with names but no descriptions are easy for competitors to steal. For companies discovering and using new and potentially improved strains, neglecting making intellectual property claims could rebound on them, as their competitors will be free to use these same strains.
In addition, companies are relying less on selling a product on its “mysterious” qualities, because with improvements in genetic typing technology competitors can now decipher the genetic fingerprint of products using their own in-house methods.
As a result, it is now more important than ever that manufacturers protect their production processes through patents. Clear-cut reproducible identification of microorganisms is becomingly increasingly important, and DNA fingerprints are ideal as supplementary information in patent application and variety registration.
Fingerprinting Techniques
Various DNA-based methods have been developed during the past decade for the identification and typing of microorganisms. The great advantage of DNA technologies is that they are very specific and able to generate a DNA profile of every individual human, animal, or microorganism. For example, in forensics, DNA profiles are used as biological evidence.
Three approaches to generate DNA profiles or DNA fingerprints are used. These methods rely on the detection of DNA polymorphism between species or individuals.
In the first method, isolated DNA is digested with restriction enzymes and the fragments are size fractionated by electrophoresis. Depending on the restriction enzyme used, micro or macrorestriction fragments are obtained, and after fractionation a specific pattern of fragments—a DNA fingerprint—is generated. Differences in patterns between species determine the level of identity or divergency.
In the second method, amplification by polymerase chain reaction (PCR) is used. Specific or randomly distributed regions are amplified by PCR, and sequence polymorphism of the fragments generated will subsequently be discovered after electrophoretic analysis. Usually, multiple amplicons (PCR fragments) are formed, and again a specific pattern or bar code is generated. Genetic differences between species result in different PCR fingerprints.
In the third method, DNA sequence analysis is used for identification. Several defined regions are amplified by PCR and subsequently sequenced. The various sequences are aligned to detect nucleotide differences. These analyses generate a multi-locus sequence database with very detailed genetic information.
Currently, the following genetic fingerprinting techniques are being used for typing microorganisms:
• Pulsed Field Gel Electrophoresis (PFGE). This is considered the “gold standard” of molecular typing methods in the field of microbiology. The embedded bacteria are subjected to in-situ detergent-enzyme lysis, and DNA is digested with an infrequently cutting restriction enzyme. The digested bacterial plugs are analyzed by agarose gel electrophoresis, allowing separation of very-large-molecular-weight-length DNA fragments ranging from 10 to 800 kilobases (kb).
This method, however, has disadvantages. It displays 10–15 DNA fragments (10 data points) per microbial sample, which is sometimes not sufficient to discriminate at the strain level. Differences between isolates are due to polymorphism in the recognition sites used or to large genomic rearrangements. Moreover, interlaboratory comparison of the method is very hard to achieve. PFGE does not yet lend itself to standardization among research centers.
--- PAGE BREAK ---
• Repetitive Sequence-Based Polymerase Chain Reaction (Rep-PCR). Bacterial genomes harbor various repetitive DNA elements such as repetitive extragenic palindromic elements (REP), enterobacterial repetitive intergenic consensus (ERIC) sequences, and BOX. PCR primers based on REP, ERIC and BOX sequences have already been designed for a large set of microorganisms.
The technique is easy to perform and can be applied to a large number of isolates. The disadvantage of this technique is that it surveys repetitive sequences only and does not give a global, whole-genome view of polymorphism throughout the complete genome. On the average, 10–15 PCR fragments per strain are obtained for comparison.
• Ribotyping. Among the typing methods used, automated ribotyping using the Riboprinter microbial characterization system (Qualicon, Warwick, UK) is the only typing method that has been almost completely automated, enabling easy comparison between strains. Genomic DNA is digested with restriction enzymes recognizing six base pairs and subsequently blotted. A large DNA probe consisting of the rRNA region of Escherichia coli is employed. On the average, 5–10 fragments varying in size between 2 and 20 kb are visualized.
The disadvantage of this method is that typing is based on a limited number of fragments derived from the rDNA region of the bacterium. Characterization is possible at the subspecies level but sometimes not at the strain level.
• Multi Locus Sequencing Typing (MLST). In this technique, bacterial isolates are characterized on the basis of the sequences of about 450-base-pair (bp) internal fragments of seven housekeeping genes. For each gene fragment, the different sequences are assigned to distinct alleles, and each isolate is defined by a specific seven-digit allele combination. As there are many alleles at each of the seven loci, isolates are highly unlikely to have identical allelic profiles by chance.
A great advantage of this method is that sequence data can be readily compared between laboratories and in different studies. Another advantage is that the loci encoding housekeeping genes are derived from different parts of the genome. However, one typing experiment requires the exact sequence determination of seven loci, rendering the method cost-effective but expensive.
• Minisatellites. Also known as variable number of tandem repeats (VNTRs), minisatellites provide a source of informative markers for identification purposes. They are present not only in humans—some of them are still used for paternity analyses and forensics—but also in a broad variety of microorganisms. Usually tandem repeats with repeat units of at least 9 bp are evaluated. The repeat unit is repeated at least six times spanning up to hundreds of base pairs. Moreover, microorganisms often harbor homopolymeric tracts and VNTRs with a unit length between 2 and 8 bp that can be applied for typing as well.
Minisatellites are easily identified from genome sequence data, and differences between bacteria are exemplified by PCR fragment sizes. Minisatellites are scattered throughout the genome and not limited to certain regions of the genome. Disadvantages of this technique are that the number of minisatellites can vary extensively among bacteria and not all tandem repeats are polymorphic. Hence the applicability is limited.
• DNA Chips. Also called DNA arrays, DNA chips are ordered arrays of oligonucleotides immobilized on an inorganic substrate. They provide new opportunities for assessing genetic diversity. This method relies on hybridization of isolated microbial DNA to large sets of oligonucleotides or DNA fragments present at a precise location on a miniaturized inorganic substrate (glass slide). This probe-based array strategy is now being investigated for bacteriological testing by various groups.
The number of oligonucleotides spotted on a chip can vary from several hundred to almost a million. The oligonucleotides can be derived from every region of the genome, and point mutations (single nucleotide polymorphisms) are targeted as well. The first chips will be commercially available in the near future for large-scale testing. Disadvantages of the method may be the cost of a chip, which can only be used once, and the need to purchase the equipment for hybridization and analysis.
--- PAGE BREAK ---
• AFLP®. Patented by Keygene NV, AFLP—an acronym for selective amplification of restriction enzyme fragments—is one of the most precise DNA typing technologies currently available for use in microbial detection and characterization (Vos et al, 1995; Savelkoul et al., 1999). It can distinguish among closely related subspecies of microorganisms, including bacteria, yeast, and fungi. It has been extensively used mostly in plant breeding, but there is a growing demand for such sophisticated technologies in the food and beverage area. A nearly 10-fold increase in AFLP citations over the past five years reflects the technique’s wide acceptance as a useful DNA fingerprinting technique in the plant science area and its increasing use for the identification of microorganisms in other sectors.
This PCR-based technology is insensitive to DNA concentration, and a broad range of PCR conditions can be applied. Simple as well as complex DNAs can be analyzed, rendering it a very universal tool for characterization of microorganisms (Fig. 1). A great advantage is that no prior sequence or any other information is required to fingerprint a microorganism of interest. Moreover, a large number of loci scattered throughout the genome can be analyzed simultaneously.
In an AFLP reaction, subsets of genomic DNA fragments are amplified. The key is the use of selective nucleotides, enabling a complete scan of the genome of interest. AFLP displays more fragments than other fingerprinting techniques, and these fragments are gathered from the whole genome.
• Variation in AFLP fingerprints is based on mutations in the recognition sites used, the use of selective nucleotides, and the length of the restriction fragments that can vary due to insertions or deletions.
Different enzyme and/or primer combinations produce AFLP patterns of different complexity. By selecting the optimal enzyme and primer combinations for every microorganism or set of microorganisms, outstanding resolving power can be obtained that exceeds that of other typing technologies.
Table 1 compares AFLP with other genetic typing methods used in food/beverage fermentations. As shown, the technique either matches or outperforms other methods on many parameters.
How the AFLP Technique Works
The AFLP concept is illustrated in Fig. 2. Genomic DNA is digested with two restriction enzymes, either with a frequent cutter (4 bp recognition) and a rare cutter (recognizing 6 nucleotides) or with two frequent cutter enzymes. For bacteria, yeast, and fungi, use of two 4-cutter enzymes enables a higher coverage to detect as much polymorphism as possible in a single reaction. Adapters are ligated to these restriction sites in such a way that the original restriction site is not restored. A subsequent preamplification uses PCR primers that anneal to the adapter sequences without any selective nucleotides.
A second amplification reaction is performed, having at the 3' ends of the PCR primers an extension of one or two selective nucleotides, generating 30–50 amplification products in the size range of 75–600 nucleotides. For bacteria, three selective nucleotides are used, whereas for fungi and yeast two selective nucleotides at either side of the fragments are used.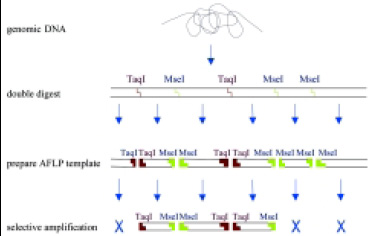
The amplified products generated are separated by gel electrophoresis, using radioactive, fluorescent, or infrared labels. Electrophoregrams are imported into gel analysis software to convert the AFLP patterns into digital images to determine the presence or absence of a fragment and to detect heterozygosity as well. This software also performs an accurate measurement of fragment lengths, a prerequisite for setting up bacterium-, yeast-, or fungus-specific AFLP fingerprint databases for future reference and interlaboratory comparison. Fig. 3 is an example of an AFLP fingerprint, showing an analysis of 23 bacterial strains involved in manufacturing of a dairy product. Very distinct differences (mutations) characteristic for a particular strain can be detected.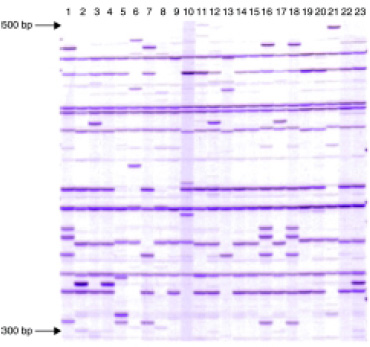
The main advantages of AFLP in genetic typing are that it is highly reproducible, easy to use, highly capable of discriminating microbial strains, does not require prior knowledge of the genome sequence, is very flexible, and is amenable to high throughput application. Because of its advantages over the other techniques discussed above, it is expected to find increased use in the food and beverage industries. 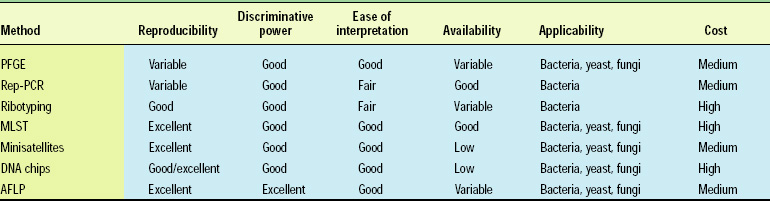
by Dian Pouwels and Guus Simons
Author Pouwels is Marketing Manager and author Simons, a Professional Member of IFT, is Program Manager, Microbial Genomes, at Keygene N.V., P.O. Box 216, 6700 AE Wageningen, The Netherlands. Send reprint requests to author Simons.
References
Savelkoul, P., Aarts, H., Haas, J. de., Dijkshoorn, L., Duim, B., Otsen, M., Rademaker, J., Schouls, L., and Lenstra, J. 1999. Amplified-fragment length polymorphism analysis: The state of the art. J. Clin. Microbiol. 37: 3083-3091.
Vos, P., Hogers, R., Bleeker, M., Reijans, M., Lee, T. van de., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M,. and Zabeau, M. 1995. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407-4414.
