Analyzing for Microbial Contaminants
LABORATORY
Analysis of foods for various microbes that can cause foodborne illness continues to be a major area of interest. This month’s column will discuss some of the events and advances that have occurred since my previous column on microbial testing, “Testing for Salmonella,” which appeared in the October 2008 issue of Food Technology.
Outbreaks and Contamination Continue
Various foodborne illness outbreaks and food recalls because of actual or potential bacterial contamination have occurred during the year.
Early this year, many consumers were sickened and nine died from an outbreak of Salmonella enterica serovar Typhimurium (referred to as Salmonella Typhimurium) in peanut butter and peanut butter products, resulting in the largest food recall in U.S. history. In March, one of the largest pistachio producers in the United States recalled its pistachio products because of possible contamination with Salmonella. An outbreak of food poisoning in April was linked to contamination of alfalfa sprouts with Salmonella enterica serovar Saintpaul (Salmonella Saintpaul), apparently from contamination of seeds. In May and June, an outbreak of Escherichia coli O157:H7 infections was linked to consumption of raw refrigerated cookie dough, which led to a major recall. Also in May, June, and July, beef and ground beef were recalled because of possible contamination with E. coli O157.H7, and in August, ground beef was recalled because of foodborne illness outbreaks linked to Salmonella enterica serovar Newport (Salmonella Newport).
The Food and Drug Administration made several “draft guidance for industry” documents, including one in June on “Measures to Address the Risk for Contamination by Salmonella Species in Food Containing a Pistachio-Derived Product as an Ingredient” and three in August to “minimize microbial food safety hazards” of tomatoes, melons, and leafy greens.
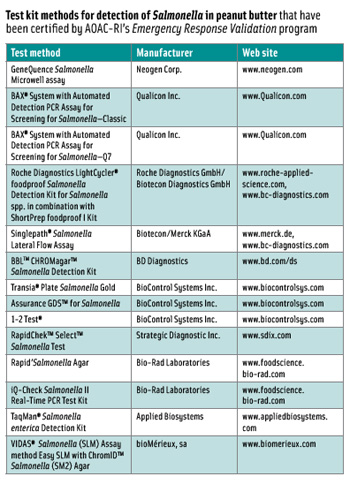
Validating Test Kits for Quick Response
Because of the outbreak of foodborne illness linked to Salmonella contamination of peanut butter, AOAC International’s subsidiary AOAC Research Institute, Gaithersburg, Md., launched a new program, Emergency Response ValidationSM (ERV), designed to respond immediately to emerging food contamination crises by rapidly evaluating test kits for specific bacteria in specific foods once a crisis is identified.
AOAC-RI had been certifying microbial test methods for many years via its Performance-Tested MethodsSM (PTM) validated methods program. The ERV program takes advantage of the organization’s existing pool of PTMs and AOAC Official Methods of AnalysisSM. All of these methods had been extensively evaluated but may not have been evaluated for the specific contaminant and food type associated with the emerging contamination crisis. The ERV program is designed to evaluate these previously AOAC approved methods for the specific contaminant and food type causing the crisis, in this case S. Typhimurum in peanut butter products.
--- PAGE BREAK ---
According to Zerlinde Johnson ([email protected]), AOAC-RI Technical Program Manager, eight companies submitted 15 test kit methods for the evaluation. Q Laboratories Inc., an independent laboratory located in Cincinnati, Ohio, conducted the evaluation. The results were compared to results obtained by the FDA Bacteriological Analytical Manual reference method for the detection of Salmonella. Regulatory agencies, such as FDA, Health Canada, and the Canadian Food Inspection Agency, also participated and received samples to evaluate their reference methods.
In February and March, the test kit methods were evaluated using blind-coded, randomized samples (20 samples for the low level, 20 for the high level, and 5 blank control samples). As a result of inherent variations in the distribution of Salmonella in peanut butter, however, the results were mixed—some methods met the acceptance criteria, while others were slightly below. Since AOAC-RI determined that most of the data did not satisfy the fractional recovery requirement and/or did not perform as well as or better than the reference method, a second round of testing was conducted in May.
The second trial utilized an improved quality assurance plan for the preparation of samples inoculated at very low levels. It addressed issues of homogeneity and microbial die-off, both of which can be a problem when inoculation levels are extremely low (less than 1 cfu/25 g) with a food that requires a long equilibration period prior to shipping. Results from the second trial showed that test kits provided satisfactory data in meeting the fractional recovery acceptance criteria for one level. Fourteen of the 15 were approved for the detection of S. Typhimurium in peanut butter and are listed in the accompanying table. The complete list of all PTMs is available at www.aoac.org/testkits/testedmethods.html.
Johnson said that the certification reports would be published in the September/October 2009 issue of AOAC’s Inside Lab Management magazine, and a full report of the evaluation would be published in the November/December 2009 issue of Journal of AOAC International. She said that as a result of lessons learned during the evaluation, AOAC-RI is revising its validation protocol to provide for more efficient future validations.
Biosensor Being Developed
Arun Bhunia ([email protected]), Professor of Food Microbiology at Purdue University, and coworkers have been developing a biosensor to identify strains of pathogenic bacteria in foods. They began working on the biochip project in 2000 and have reported on various stages of the research. The most recent paper, “Targeted Capture of Pathogenic Bacteria Using a Mammalian Cell Receptor Coupled with Dielectrophoresis on a Biochip,” by O.K. Koo, et al., appeared in Analytical Chemistry, Vol. 81, pp. 3094-3101. The researchers have been working initially with Listeria monocytogenes but plan to expand the work to include Salmonella, E. coli, and other bacteria.
--- PAGE BREAK ---
Bhunia—who received the Institute of Food Technologists’ 2009 Research and Development Award for his work on early detection of foodborne pathogens to reduce the risk of foodborne illness outbreaks—said that the biosensor is a microfluidic biochip, about the size of a postage stamp, onto which a pathogen-specific antibody or similar receptor molecule is immobilized. When bacteria in a fluid are passed through the biochip, the bacteria bind to the receptor and grow inside the chip. Electrodes measure the change in impedance as the bacteria grow. They are currently using polymerase chain reaction (PCR) on-chip for further identification.
Bhunia said that they previously used antibodies as the receptor molecule but are now using the protein that L. monocytogenes uses to infect human cells. The biochip consists of heat shock protein 60 (Hsp60), a eukaryotic mitochondrial chaperon protein, immobilized onto the surface of streptavidin-coated silicon dioxide as a receptor for Listeria adhesion protein during Listeria infection. Use of this specific protein enables the system to differentiate between pathogenic and nonpathogenic species, he said, making the biochip more sensitive and specific. He uses commercially available antibodies and proteins, as well as ones he has developed.
He said that after expanding the work to other bacteria, future work will involve trying to do on-chip PCR for other bacteria, including Salmonella and E. coli O157:H7. He would then have a lab-on-a-chip, using standard reagents. Current PCR methods for Listeria and other pathogens generally take 1–10 days to obtain results, he said, but the biochips take less than 12 hr.
Laser Instrument Identifies Strains
Bhunia and his coworkers have also developed a laser light-scattering instrument that can detect and identify strains of pathogens in food with high accuracy (specificity).The instrument was developed with USDA funding by Bhunia and coworkers in the Dept. of Food Science and colleagues in the Dept of Mechanical Engineering and the Dept of Basic Medical Science at Purdue. After a sample—food, water, air, etc.—is added to agar on a Petri dish and incubated, the dish is placed into the instrument, and a laser selects each colony and produces a scatter pattern that can be used as a signature or fingerprint of the specific bacterium, both genus and species and even at serovar levels.
Each species, Bhunia said, has a specific pattern, and they already have extensive libraries of thousands of patterns for several foodborne pathogens, including Listeria, E. coli, Vibrio, and Salmonella. The instrument can scan 100 colonies in five minutes, he said, compared to PCR, which requires 1–3 hr for each colony. The instrument scans each colony and compares the scatter pattern to the image library. The user can look for a particular species or simply determine what’s present.
The system was described in a 2009 paper, “Label-Free Detection of Multiple Bacterial Pathogens Using Light-Scattering Sensor” by P.P. Banada et al. in Biosensors and Bioelectronics, Vol. 24, pp. 1685–1692.
Although the system requires incubation of the sample for 12–24 hr, Bhunia said, one of its benefits is that it doesn’t destroy the colony, allowing it to be used for verification or further testing.
--- PAGE BREAK ---
Advanced Bioimaging Systems (http://advancedbioimagingsystems.com) licensed the Purdue technology and has produced several prototypes. Brian Straight ([email protected]), Chief Executive Officer, said that the first instrument (named BARDOT, for Bacterial Rapid Detection Using Optical Scanning Technology) was delivered to USDA’s Agricultural Research Service in Pennsylvania in July for use in building more comprehensive pathogenic pattern libraries with the instrument.
The second will be delivered soon to USDA in California for use in studying contamination of leafy green vegetables, essentially to produce a general microbiological assay of conditions such as soil, root, and leaf characteristics and standard microbiological flora. This second instrument, named the BARDOT LS-102A, he said, is one-third the size and weight of the original prototype, with a simplified and more rugged construction. It has a very simple interface, and users can be trained within a half hour. Straight added that the instrument is production-ready and the company welcomes inquiries from potential distributors and partners.
Other Developments in Brief
• DuPont Qualicon (www.qualicon.com) announced that two BAX® System 24E assays have received certification from the AOAC Research Institute as next-day methods for detecting Listeria in food and environmental samples. Validation studies on hot dogs, spinach, cooked shrimp, queso fresco cheese, and stainless steel showed that the systems performed faster and as well as or better than traditional culture methods. The company also announced that a new BAX® System assay can be used by seafood processors and government laboratories to detect Vibrio in less than 24 hr. The BAX System Real-Time PCR Assay for Vibrio detects even low levels of three distinct species—Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus—from the same sample. Tested on shrimp, tuna, oysters, scallops and crab, the system is said to deliver reliable, differentiated results in less than one day, with results equivalent to or better than the reference culture method, which typically takes 3–5 days. BAX is an automated system that uses leading-edge technology, including PCR assays, tableted reagents, and optimized media, to detect a variety of other bacteria, as well, including Salmonella, L. monocytogenes, E. coli O157:H7, Enterobacter sakazakii, Campylobacter, and Staphylococcus aureus, and yeast and mold.
• Meridian Bioscience Inc. (www.meridianbioscience.com) announced that it has received FDA clearance for its new Premier™ Campy rapid EIA test for optimized detection of Campylobacter infection, the most commonly diagnosed bacteria for foodborne illness in the U.S. Approximately 20 million tests are conducted each year in the U.S. to detect the illness, which is usually caused by poorly cooked poultry. The company has also received FDA clearance to market ImmunoCard Stat!® EHEC, a new test for the diagnosis of E. coli infection. Kit detects all Shiga-toxin producing E. coli and is said to be the first rapid (20-min) E. coli diagnostic that differentiates between Toxin 1 & Toxin 2.
• ADRIA Développement (www.adria.tm.fr) has issued software that can be used in conducting shelf-life studies of foods in accordance with EC regulations on microbiological criteria for foodstuffs. Sym’Previus aids in the determination of the shelf life of products using the probability approach. It can also be used to classify the products in which growth of L. monocytogenes and other foodborne pathogens can occur or will not occur during their shelf life and can also simulate thermal destruction.


