Targeting Cyclosporiasis
FOOD SAFETY AND QUALITY

Outbreaks of cyclosporiasis, a foodborne and waterborne illness caused by the intestinal parasite Cyclospora cayetanensis, have been reported in the United States since the 1990s and have been linked to various types of fresh produce, including raspberries, snow peas, mesclun lettuce, basil, and cilantro. According to the Centers for Disease Control and Prevention (CDC), there were 33 reported foodborne outbreaks of cyclosporiasis in the United States between 2000 and 2016 and several outbreaks in 2018, including two multistate outbreaks with 2,299 laboratory-confirmed cases. Of these two, one was associated with packages of dip and pre-cut broccoli, cauliflower, and carrots, and the second was associated with prepackaged salads containing carrots, romaine lettuce, and other leafy greens. Although the U.S. Food and Drug Administration (FDA) did not identify a single source or potential point of contamination for either outbreak, romaine lettuce was considered to be the likely source of the second outbreak. These two outbreaks accounted for less than half of all domestically acquired Cyclospora cases reported to the CDC in 2018. Smaller clusters of illness in 2018 were also identified and linked to basil and cilantro.
The FDA has increased its sampling of food commodities to monitor for the presence of foodborne pathogens, including screening for Cyclospora. The agency has been working closely with the National Association of State Departments of Agriculture to train federal and state regulators who will conduct inspections and to provide training opportunities for farmers to help them implement the Food Safety Modernization Act’s produce safety rule, which establishes minimum standards for the safe growing, harvesting, packing, and holding of fruits and vegetables for human consumption.
C. cayetanensis is a microscopic protozoan parasite. Infected persons shed immature (noninfective) oocysts in their feces, and it takes eight to 14 days (10 days on average) for these oocysts to sporulate and become infectious. If ingested from contaminated food or water, the sporulated oocysts infect the host, causing diarrhea and other symptoms. The oocysts are highly resistant to disinfectants and sanitizing methods commonly used in the food industry.
The infection is spread by ingestion of food or water contaminated with the sporulated oocysts, not by person-to-person contact. The infectious dose is not known. The time between becoming infected and becoming sick is usually about one week. Some people have no symptoms, and symptoms may go away and then return. If not treated, the illness may last from a few days to a month or longer. C. cayetanensis is considered the only species of Cyclospora that causes cyclosporiasis in humans.
 Cyclospora Detection
Cyclospora Detection
C. cayetanensis cannot be cultured, so cyclosporiasis is diagnosed by examining stool samples under the microscope. If a modified acid-fast staining technique is used, oocysts typically appear light pink to deep red or purple or unstained with a wrinkled appearance. If a modified safranin staining technique is used, oocysts uniformly appear bright reddish orange. If viewed under a fluorescence microscope, oocysts, which are autofluorescent, appear blue or green against a black background. If suspect oocysts are found by any of these methods, bright-field, phase-contrast, or differential interference contrast microscopy can be used to confirm that the structures have the characteristic morphologic features of Cyclospora oocysts. Identification by microscopy is challenging because of lack of sensitivity to detect low concentrations of oocysts in food matrices, inability to discriminate Cyclospora species that are morphologically identical to C. cayetanensis, and low concentrations of oocysts in stool samples. Molecular diagnostic methods, such as polymerase chain reaction (PCR) analysis, can be used to identify the parasite’s DNA in the stool.
The FDA in 2004 included a method for molecular identification of C. cayetanensis in fresh produce in Chapter 19a of its Bacteriological Analytical Manual (BAM), which details the agency’s preferred laboratory techniques for the detection of pathogens and microbial toxins in food and cosmetic products. The method involved washing the produce, isolating the DNA from the wash, and identification by nested PCR. The method became impractical when certain required supplies were no longer commercially available.
In late 2014, the FDA’s Center for Food Safety and Applied Nutrition began establishing a Foodborne Parasitology Research Program under the leadership of senior parasitologist Alexandre J. da Silva to address major gaps in foodborne parasitology. The program had a primary focus on creating a new, more sensitive method to detect and characterize C. cayetanensis in food and environmental samples. Da Silva and research microbiologist Helen Murphy developed a method that could be used for regulatory testing of fresh produce, including leafy greens such as lettuces, cilantro, and basil; soft fruit such as raspberries, blackberries, and strawberries; and whole vegetables such as beans and peas. The new method, which uses real-time PCR (also called quantitative PCR or qPCR), was validated in 2016 by Murphy, da Silva, and Sonia Almeria for cilantro and raspberries in a multi-laboratory study led by Murphy and was adopted in July 2017 as BAM chapter 19b, replacing chapter 19a. It was extended to include shredded carrots, parsley, and basil in 2017 and packaged pre-cut romaine lettuce salad in 2018.
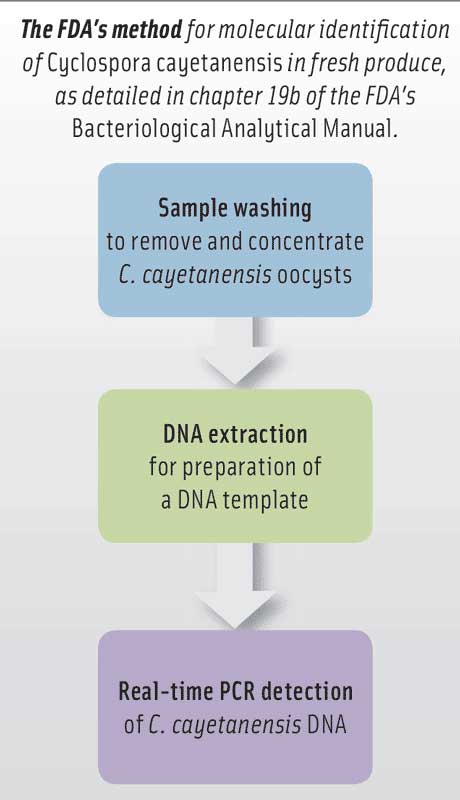 The method in chapter 19b comprises three main steps: sample washing with Alconox detergent to remove and concentrate C. cayetanensis oocysts from produce; DNA extraction using MP Biomedical’s FastDNA SPIN Kit for Soil and FastPrep-24 5G Homogenizer for preparation of a DNA template; and real-time PCR detection of C. cayetanensis DNA, using the Applied Biosystems 7500 Fast Real-Time PCR System and TaqMan probes by Fisher Scientific. Da Silva said that the method offers increased sensitivity, specificity, and throughput; significantly reduces analytical time; and minimizes laboratory environment contamination commonly associated with conventional PCR.
The method in chapter 19b comprises three main steps: sample washing with Alconox detergent to remove and concentrate C. cayetanensis oocysts from produce; DNA extraction using MP Biomedical’s FastDNA SPIN Kit for Soil and FastPrep-24 5G Homogenizer for preparation of a DNA template; and real-time PCR detection of C. cayetanensis DNA, using the Applied Biosystems 7500 Fast Real-Time PCR System and TaqMan probes by Fisher Scientific. Da Silva said that the method offers increased sensitivity, specificity, and throughput; significantly reduces analytical time; and minimizes laboratory environment contamination commonly associated with conventional PCR.
The DNA extraction step of the chapter 19b method uses the MP Biomedical kit, da Silva said, because it is very efficient in disrupting parasites’ oocysts and has been used for several years by parasitology groups worldwide. While no other DNA extraction technique is recommended in the method, Qiagen and other companies also supply DNA extraction kits. In addition, the method requires specific reagents for real-time PCR detection. Verification studies comparing different real-time PCR platforms need to be conducted, da Silva added, to determine whether they have the same performance characteristics as the validated Applied Biosystems platform.
There are not a lot of options for detection of C. cayetanensis in food, da Silva said, but other diagnostic platforms exist for clinical diagnosis of cyclosporiasis. For example, the BioFire FilmArray Gastrointestinal Panel, a multiplex platform capable of detecting 22 intestinal pathogens, including bacteria, viruses, and C. cayetanensis and three other protozoan parasites, has been cleared by the FDA for in vitro clinical diagnosis. Also, Primerdesign’s Genesig real-time PCR kit provides detection and simultaneous quantification of C. cayetanensis and other pathogens. Personnel from da Silva’s group, the FDA’s Office of Regulatory Affairs, and other FDA offices held workshops in 2018 to train analysts from FDA field laboratories on the method, and two more training workshops are scheduled for 2019. Five field laboratories are now fully proficient in the chapter 19b method, and two more are expected to have C. cayetanensis testing capabilities by spring 2019.
Da Silva said that his group has modified the chapter 19b method to detect C. cayetanensis in water. The method relies on ultrafiltration, using hollow-fiber filters to concentrate the parasite oocysts and other types of pathogens (bacterial and viral) from large volumes of agricultural water. This is crucial, he said, because the parasite cannot be propagated in vivo or in vitro from food, clinical, or environmental samples, and the low concentration of oocysts in water requires large amounts of sample to be screened. The method uses the same DNA extraction procedure and real-time PCR assay that is described in chapter 19b with minor modifications. This type of robust filtration and concentration should play an important role in investigations targeted at identifying potential sources of produce contamination, he said. The method has been single-lab validated, and a multi-lab validation study involving multiple FDA centers and offices began in September 2018.
Determining Genetic Makeup
Before 2014, there was no way of comparing the strain of C. cayetanensis that infected a person with a strain of C. cayetanensis found in a food source. Scientists from the CDC, the FDA, state public health departments, and academia are working to develop and validate molecular typing methods that could distinguish among different strains of C. cayetanensis. Those methods would facilitate linking cases of cyclosporiasis to each other and to food vehicles and their sources and could in the future be applied for the genotyping of C. cayetanensis from food and environmental samples. Da Silva’s group is working on those methods as well and has developed laboratory methods and bioinformatic approaches to sequence, assemble, and analyze Cyclospora genomes from oocysts isolated from fecal samples. There are 16 whole genome assemblies in the Cyclospora cayetanensis GenomeTrakr BioProject, da Silva said, and the creation of this BioProject represented a major step toward building a network that researchers worldwide could take advantage of and contribute C. cayetanensis genome data to. Da Silva said that his group has already sequenced approximately 20 additional genomes and 40 organellar genomes (mitochondria and apicoplast genomes), and the data accumulated is providing enough information to start working on molecular epidemiology approaches. With the preliminary genomic data obtained over the past two years from the limited number of samples analyzed so far, he said, it was possible to find regions in the C. cayetanensis genome that have a certain level of polymorphism that could be used to distinguish distinct types of C. cayetanensis. However, he added, scientists still need to analyze a much larger and diverse sample set to better understand how to use these genetic markers to create a robust molecular epidemiology method for source tracking.
The CDC has a website called DPDx, which provides concise reviews on parasites and parasitic diseases; an image library; and a review of recommended procedures for collecting, shipping, processing, and examining biological specimens. Via the website, laboratory personnel and health professionals can ask questions and send digital images of specimens for free expedited review and consultation with DPDx staff.
Challenges Ahead
Da Silva said that Cyclospora is peculiar in many ways and thus presents major challenges. One is that the oocyst load is low in both water and contaminated produce and thus requires very sensitive methods for detection and characterization. Another is the absence of methods to propagate the parasite in vivo or in vitro; this alone poses an enormous scientific challenge that slows the development of processes to inactivate the parasite in irrigation water or on fresh produce. Yet another is sequencing the genome. The Cyclospora genome is approximately 10 times larger than common bacterial pathogens, and sequencing such a large genome from stool samples is very difficult. Therefore, assembling whole C. cayetanensis genomes also relies on refined bioinformatic pipelines (i.e., a metagenomics approach). Despite all these challenges, he said, considerable progress is being made.
Other challenges are related to the diversity and dispersion of C. cayetanensis and other species of Cyclospora in the environment. Improving the knowledge in this area, da Silva said, will help develop mitigation strategies to prevent food contamination. To date, C. cayetanensis is considered the only species of this genus that causes human infections, but several species of Cyclospora occur in animals and can be morphologically identical to C. cayetanensis; however, this does not mean that they are the same species. More studies are needed to address the knowledge gaps about Cyclospora, da Silva said.
 Neil H. Mermelstein, IFT Fellow,
Neil H. Mermelstein, IFT Fellow,
Editor Emeritus of Food Technology
[email protected]


