Targeting Non-O157 E. coli Serotypes
FOOD SAFETY & QUALITY
Scientists at DuPont Qualicon (www.qualicon.com) and the U.S. Dept. of Agriculture’s Agricultural Research Service are working together to develop a rapid method for detecting and identifying hard-to-detect strains of Escherichia coli that have been causing increased instances of foodborne illness around the world.
Rising Concern
Some strains of E. coli produce Shiga toxin, which can result in foodborne illness ranging from diarrhea to hemorrhagic colitis and hemolytic uremic syndrome, which can be deadly. The most commonly identified Shiga toxin-producing E. coli (STEC) serotype is O157:H7, but there are more than 200 other STEC serotypes that have been associated with human illness. O157 is referred to as the serogroup, and when the H type is also known (e.g., H7), O157:H7 is referred to as the E. coli serotype.
The Centers for Disease Control and Prevention (CDC) has estimated that non-O157 STEC bacteria are responsible for 36,000 illnesses and 30 deaths each year in the United States. The majority of these infections have been associated with six serogroups: O26, O45, O103, O111, O121, and O145. The most recent out-break was in spring 2010, when the O145 serogroup was found in shredded romaine lettuce.
Since the reporting of all STEC illnesses to CDC began in 2001, instances of non-O157 STEC illnesses have steadily increased. USDA research microbiologist/research leader Pina Fratamico said that in the past 25 years there have been at least 30 outbreaks associated with non-O157 STEC serotypes in the United States. Foods involved have included pasteurized milk, punch, lettuce, pasteurized cider, berries, and lettuce; ground beef has also been suspected. In addition, outbreaks have been associated with drinking and recreational water, animal contact, and person-to-person transmission.
The non-O157 STEC serotypes are not as well understood as the O157:H7 serotype partly because such outbreaks are rarely identified, many infected people do not seek medical care, and many clinical laboratories do not test for STEC infection since this group of bacteria are more difficult to identify than E. coli O157.
To prevent human infections with non-O157 STECs, Fratamico said, it is important to have reliable methods for detecting these pathogens in food. The test being developed with DuPont Qualicon will be rapid and simple to perform and will be useful for screening the top six non-O157 STEC serogroups in food.
--- PAGE BREAK ---
Testing for E. coli O157:H7
Diagnosis of O157 STEC illnesses involves taking a stool specimen, testing for Shiga toxins, isolating a colony on sorbitol-MacConkey agar, agglutinating with O157 antiserum, and then conducting pulsedfield gel electrophoresis. Food testing requires a similar method, which is time-consuming and requires trained microbiologists. Consequently, DuPont Qualicon technical and regulatory specialist Amy Smith said, the company developed its BAX® System to bring high-tech capabilities to food laboratories that may not have highly skilled scientists on staff.
The system uses simplified sample preparation, tableted reagents, and polymerase chain reaction (PCR) assays to detect specific bacteria. It was introduced for detection of Salmonella in 1995, and the company has since developed applications for detection of many other pathogens (see table below).
In 2005, USDA’s Food Safety and Inspection Service (FSIS) began using the BAX System E. coli O157:H7 MP assay to monitor meat and poultry. A few years later, DuPont Qualicon began working with USDA scientists to develop a new assay that used real-time technology to detect E. coli O157:H7. The BAX System Real-Time PCR Assay for E. coli O157:H7 was introduced in 2009 and has been certified by the AOAC Research Institute as a Performance Tested MethodSM for detection of E. coli O157:H7 in raw ground beef, beef trim, lettuce, and spinach.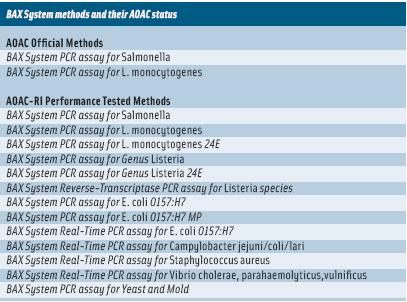
The assay uses PCR to amplify (replicate) specific sequences of bacterial DNA that are unique to E. coli O157:H7. The company simplified the PCR process, Smith said, by combining the required reagents into a dry tablet that is hydrated with a prepared sample and processed in the BAX System instrument. The instrument amplifies the targeted DNA fragments through a series of heating and cooling cycles. During each cycle, the instrument measures the change in fluorescence of the Scorpion® probes (see sidebar) and translates that information into a visual indication of presence or absence of the targeted organism within an hour of loading samples. The time to results, Smith said, can be as quick as the same day as sample collection; other methods can take days.
Developing an Assay for Non-O157 Serotypes
Detection methods for non-O157 STEC serotypes are extremely limited, Smith said, because these serotypes are very hard to identify. Some grow much more easily than others, and there are few outstanding characteristics to differentiate them from one another or from background flora.
Fratamico said that the current testing for STEC serotypes relies on culture-based methods, namely enrichment followed by plating onto selective agar, confirmation of presumptive colonies by biochemical tests, and serotyping to identify the O somatic and H flagellar antigens. PCR-based methods targeting Shiga toxin genes and other virulence genes have also been used to screen enrichments for STEC, followed by plating onto selective agars and confirmation of presumptive colonies.
--- PAGE BREAK ---
Smith said that nonculture methods such as enzyme immunoassays and PCR are used to detect non-O157:H7 STEC serotypes, but there is little standardization or uniformity to their use. Thus, there is a need for an easy, rapid method that can be used by diagnostic laboratories as well as food and regulatory laboratories. Fratamico added that enzyme immunoassays will only determine whether the sample contains one or more strains that produce Shiga toxins but will not provide information on the serotype. To do that requires isolating the organism and performing serotyping.
The scientists at DuPont Qualicon in Wilmington, Del., and USDA’s Eastern Regional Research Center in Wyndmoor, Pa., are working together to design a PCR assay and determine test conditions to detect non-O157 STEC serotypes. DuPont Qualicon will incorporate the reagents needed into tablets, as in other BAX System kits, and USDA scientists will test them. Fratamico said that the test will target important STEC virulence genes as well as genes in the O antigen gene clusters of important STEC serogroups, thus identifying the O group.
“USDA continually looks for opportunities to collaborate in ways that will expedite research to assist regulatory agencies and move technologies into the marketplace,” Fratamico said. “This collaborative project to develop a discriminating STEC test is a good fit with our mission.”
There are three major areas of focus in the design of this assay, Fratamico said: (1) enrichment of the six serogroups, making sure that the method can allow propagation of all six non-O157 STEC serogroups to detectable levels; (2) real-time PCR detection; and (3) isolating a colony on agar medium to confirm that the non-O157 STEC serogroup is in the sample. Once a reliable test method has been developed, she said, FSIS will perform a baseline study to determine which non-O157 STEC serogroups are in a beef supply and then consider whether those serogroups will be considered adulterants in meat. At present, FSIS is concerned with whether the strain belongs to one of the top six serogroups regardless of what H type they possess. However, the strains belonging to the six serogroups must possess the Shiga toxin and eae genes.
The method is in the very early stages of development and is estimated to launch in 2011, Smith said. When the company releases the method, it will be submitted to the AOAC Research Institute for Performance Tested Method certification.
--- PAGE BREAK ---
Regulatory Matters
USDA’s FSIS considers E. coli O157:H7 an adulterant in ground beef, but it’s the only STEC serotype considered as such. In October 2009, foodborne illness lawyer Bill Marler petitioned FSIS to declare all non-O157 STECs to be adulterants in ground beef, and in April 2010, U.S. Senator Kirsten E. Gillibrand (D-N.Y.) asked USDA to declare all STEC serotypes to be adulterants and to accept a petition by Safe Tables Our Priority (S.T.O.P.) to expand the definition of adulterant to include E. coli O157:H7 and the six major non-O157 STEC serotypes in any type of beef, not just ground beef or beef intended for ground beef.
FSIS replied that it cannot reach a decision about the substance of the petition until it has developed additional laboratory capacity to detect and isolate various non-O157 STEC serogroups.
In May 2010, Senator Gillibrand proposed legislation (S. 3435) that would, among other things, specifically name E. coli strains O157:H7, O26, O45, O103, O111, O121, and O145 as adulterants in beef and require USDA and beef manufacturers to test for all seven strains and dispose of meat product in which any of the strains is found. The bill has been referred to the Committee on Agriculture, Nutrition, and Forestry.
The American Meat Institute (AMI) stated that no confirmed outbreak of any of the six non-O157 strains in the bill has ever been associated with a meat product, there is no test currently available to easily detect them, and USDA experts have said in public meetings that food safety systems currently in place work equally well against O157 and non-O157 STEC serotypes.
AMI said that additional research is needed to quantify the prevalence and virulence of these additional strains in beef and to ensure not only that an effective test is readily available but also that the test could be easily obtained and quickly administered.
How the BAX System Works
After standard collection steps, a sample of food is added to enrichment media, homogenized, and incubated. The enrichment step takes as little as 8 hr for produce and 9 hr for ground beef. Next, a lysis reagent breaks the cell wall and releases the DNA. PCR tablets are hydrated with a sample of the lysate, and the mix is then processed in the BAX System instrument.
The BAX System Real-Time PCR Assay for E. coli O157:H7 uses Scorpions technology to target selected sequences of the DNA. The Scorpions consist of a primer (targeted DNA sequence) and a hair-pin-shaped probe with fluorescent and quencher dyes in close proximity. A series of heating cycles open the double-stranded DNA to form single strands, followed by cooling cycles that allow primer to attach to the targeted DNA sequence. Polymerase extends the DNA into a complementary strand. With the next heating cycle, double-stranded DNA separates and the hairpin structure of the probe unfolds. Then cooling causes the probe segment to bind to the amplified target sequence. This prevents the hairpin structure from reforming and allows the fluorescent dye to emit a signal, which is measured at the end of each cycle. As the process is repeated over and over, the fluorescent signal, if present, increases with each cycle to a level that is interpreted as positive by the instrument. If the target is not present in the sample, the primers do not bind and fluorescent signal is not released.
The results are presented—within an hour—as color-coded icons indicating presence or absence of the target. In validation studies, the BAX System has demonstrated extremely low limits of detection: one cell per sample volume of 65 g of ground beef, 375 g of beef trim, and 25 g of spinach and lettuce.
by Neil H. Mermelstein, a Fellow of IFT, is Editor Emeritus of Food Technology
[email protected]


