IFT Expert Report on Biotechnology and Foods
Introduction
The Institute of Food Technologists convened three panels of experts, consisting of IFT members and other prominent biotechnology authorities, to prepare a comprehensive scientific review of biotechnology. The report consists of four sections. The first section— Introduction— appears in this issue of Food Technology, and the next three sections—Safety, Benefits and Concerns, and Labeling—will appear in the September issue.
The use of modern biotechnology to produce foods and food ingredients is a subject of significant public interest today, at the consumer, public policy, and scientific levels. The popular press and media have reported a wide range of views on these foods and food ingredients.
To promote a meaningful public discussion of these foods and food ingredients, IFT has commissioned three expert panels to review the available scientific literature on three different, but related aspects, of these foods and food ingredients: human food safety, benefits and concerns, and labeling. The panels’ report will also discuss some of the public policy implications of the underlying science.
In keeping with the widespread usage in the popular press and media, the report uses the terms “rDNA biotechnology” and “rDNA biotechnology-derived foods” to describe the application of recombinant DNA, or rDNA, technology to the genetic alteration of plants and microorganisms, and foods made therefrom. This technology, commonly known as genetic modification or gene splicing, allows for the effective and efficient transfer of genetic material from one organism to another. Instead of cross-breeding plants for many generations or introducing mutations to introduce a desired trait—processes that are imprecise and that sometimes introduce unwanted changes—scientists can identify and insert one or more genes responsible for a particular trait into a plant or microorganism with greater precision and speed, although the current technology produces gene insertions at random locations. These transferred genes, or transgenes, do not have to come from a related species in order to be functional, and can be moved virtually at will among different living organisms.
This report focuses on rDNA biotechnology-derived foods, food ingredients, and animal feed of plant origin, and on the use of rDNA biotechnology-derived microorganisms such as yeasts and enzymes in food production. While milk from cows that have received rDNA biotechnology-derived hormones is discussed, transgenic animals resulting from the application of rDNA biotechnology techniques to animal reproduction are beyond the scope of this report. Health and medical benefits associated with rDNA biotechnology-derived plants are discussed briefly.
This first section presents background information to assist the reader in understanding rDNA biotechnology-derived foods. It will first discuss biotechnology in the broad sense and how rDNA biotechnology-derived foods are the latest step in a 10,000-year sequence of human intervention in the genetic improvement of food, then it will discuss federal regulation and oversight of rDNA biotechnology.
Overview of Biotechnology
Biotechnology in the broad sense is, in fact, not a discrete technology. It refers to a group of useful enabling techniques, including but not limited to genetic modification, that have wide application in research and commerce. Over the past several decades, such techniques have become so integrated into the practice of plant breeding and microbiology and so commingled with conventional techniques as to blur distinctions between “old” and “new.” A useful working definition of biotechnology used by several United States government agencies is the application of biological systems and organisms to the production of useful goods and services. These encompass advances in biology, genetics, and biochemistry to technical and industrial processes as different as drug development, fish farming, forestry, crop development, fermentation, and oil spill clean-up (OTA, 1984).
--- PAGE BREAK ---
Turning to food biotechnology, the history of the development of modern genetics and molecular biology, which underpins much of this technology, has been discussed and reviewed by a number of authors. Two accounts accessible to interested non-specialists are those by Grace (1997), and Watson and Tooze (1981). Historically, the key role played by deoxyribonucleic acid (DNA) in determining the mechanism of inheritance in all living organisms was first established by Avery et al. (1944), who, using S and R type pneumococci, showed that DNA from one strain of bacteria can be taken up by a different strain, hereditarily altering that second strain. This pivotal demonstration was the first description of transformation, a mechanism of gene transfer that involves the uptake and integration of isolated DNA by an organism. It is a phenomenon that is central to an understanding of rDNA biotechnology, and may even be said to mark the beginning of the concept of the new biotechnology.
Geneticists had earlier recognized that the chromosomes, linear structures composed of DNA and protein, were the vehicles of inheritance in the sense that they carried genes determining inherited characteristics. Genes were conceived of as beads on a string. Genes that encode similar functions in different organisms are called orthologs (also loosely called homologs), and genes that have the same structure in different organisms are said to have synteny (also loosely called homology). Many organisms are diploid, that is, they have two sets of chromosomes, one inherited from each parent. The pairs of chromosomes are present, in a constant and characteristic number, in all the cells of an organism.
When the cells divide, the chromosomes also divide equally, by a process called mitosis. When a diploid organism prepares for sexual reproduction by forming gametes, a reduction division, called meiosis, reduces the number of chromosomes so that each egg or sperm cell has exactly half the diploid number. At meiosis, there is a random assortment of maternally and paternally derived chromosomes, which is further complicated by exchanges between paired homologous chromosomes due to “crossing over” that takes place between chromosomes. Thus, in a sense, the genetic constitution of each gamete resembles a hand of cards dealt from a well-shuffled deck. In nature, gametes (germ cells) generally unite randomly at fertilization to restore the diploid condition. Plant breeders use this variation by selecting the best plants that result from these combinations and stabilizing them by inbreeding or propagating them vegetatively. Thus, sexual reproduction produces “recombinant” organisms, in the sense that the organisms possess DNA rearranged and combined from two separate organisms.
The task of plant and animal breeders is to select individuals that retain in a heritable way the desirable features of the parent lines. The segregation of genes with easily detected effects, such as round versus wrinkled seeds, was observed by Mendel, who first described the discrete nature of inheritance in peas.
Twentieth-century plant breeding, even before the advent of modern rDNA biotechnology methods, sought ways to take advantage of useful genes and gradually has found a wider and wider range of plant species and genera on which to draw. Breeders have long used interspecies hybridization, transferring genes between different, but related, species. Subsequently, plant geneticists found ways to perform even wider crosses between members of different genera using tissue culture techniques. Crops resulting from such wide crosses are commonly grown and marketed in the U.S. and elsewhere. They include familiar and widely used varieties of tomato, potato, corn, oat, sugar beet, bread and durum wheat, rice, and pumpkin.
Although DNA was known to play a key role in inheritance, it was not until Watson and Crick (1953) described the structure of the double-stranded DNA molecule that scientists understood how the exact replication of the DNA occurred at each cell division and how the sequence of nucleotides in the DNA molecule determined the sequence of nucleotides in messenger ribonucleic acid (mRNA) and in turn, through a triplet code, the sequence of amino acids in a protein.
--- PAGE BREAK ---
When the DNA sequence of a gene is expressed, it is transcribed to form a single-stranded mRNA molecule, which is translated to make a protein. It is now known that the instructions for programming the development of a fertilized egg cell, or zygote, into an adult organism composed of millions of cells carrying identical sets of genes are encoded in the nucleotide sequence of the DNA. This is in the form of a code based on the four nucleotides, adenine, thymine, cytosine, and guanine, which form a series of three-letter words, or codons, that specify the amino acid sequences of the many thousands of proteins that carry out the cellular functions.
Biochemists have established that the basic metabolic events in all organisms have far more in common than was previously suspected. They found that not only is DNA the universal code used by all living things, but that the central functions of all organisms are nearly identical. DNA and ribonucleic acid (RNA) replication, protein synthesis, photosynthesis, energy metabolism, and a host of other functions were found to have much in common throughout living systems. Molecular biologists soon learned to determine the sequences of genes that encoded these properties.
As more and more genes were sequenced and compared, scientists found that the products of the genes that encode similar traits in very diverse organisms are often very similar in protein sequence. It also became apparent that most genes do not have characteristics specific to the organism in which they are found. In fact, it is impossible to determine the organism from which a gene arises by inspection of the gene sequence alone, although codon usage does vary among major groups of organisms. Put another way, there is no way to identify “fish genes,” “tomato genes,” or “broccoli genes.” The uniqueness of organisms instead lies not only in the DNA sequences of their genes, but also the organization of the genes which are present, and at what time and to what extent they are expressed.
Enormous quantities of DNA have now been sequenced for a wide range of organisms. The genomes (the totality of genetic material) of several bacteria and small organisms have already been fully sequenced, and the genome sequences of higher organisms such as plants, insects, animals, and humans will soon be available. In fact, about 40 genomes are expected to have been sequenced by the end of 2000 (Lander and Weinberg, 2000). Even sequencing of the human genome is now more than 90% complete. One key observation is that, in the course of determining DNA sequences, identical genes are regularly found in organisms that are only remotely related. This observation has provided evidence that genetic transfer has occurred in nature to produce natural rDNA-containing organisms.
A discovery important to modern rDNA biotechnology techniques (Linn and Arber, 1968) was the recognition that a series of so-called “restriction enzymes,” thought to protect cells from invading viral DNA, could be used to cut the DNA at precise sites defined by the sequence of four, five, or six nucleotides at the site where the cut would be made. By using DNA ligases—enzymes that fuse together two pieces of DNA—the pieces of DNA formed by cutting DNA with restriction enzymes could be joined together into a single piece of DNA. The fragments or pieces of DNA could also come from two different organisms. Pieces of DNA from different organisms are often called “heterologous DNA” and when heterologous fragments of DNA are joined together by a ligating enzyme, the fragment of DNA is said to be a “recombinant” molecule; i.e., it recombines DNA from two heterologous sources. The word “recombinant” is used analogously to describe the recombination of DNA of the parental chromosomes that takes place during meiotic cell division.
This ability to splice together pieces of heterologous DNA means that it is possible to clone fragments of DNA by splicing them into a bacterial plasmid, a circular self-replicating DNA molecule that multiplies inside the bacterial cell when it is introduced into the bacteria by a process called transformation. If the heterologous DNA was spliced into a site on the plasmid where the DNA would have an opportunity to be transcribed to mRNA, and then translated to form a functional and active protein, its action in the cell can be detected so that the function of the cloned fragment can be identified. By this means, it is possible to produce very large numbers of copies of a known DNA fragment that can then be used to transform other organisms, such as plants and animals.
--- PAGE BREAK ---
Two methods of plant transformation are in use at the present time. One method, known as the ballistic or free DNA method, uses a gun to shoot microscopic particles of gold or tungsten into cultured plant cells. The particles are first coated with the DNA carrying the gene of interest, isolated from the bacteria in which it has been cloned. Then, these particles are accelerated by releasing a charge of helium under high pressure. A small proportion of the particles penetrate not only the plant cell wall but the nuclear membrane as well. The DNA carried by these particles can be taken up and integrated into plant chromosomes.
Although the entire nucleotide sequence of the segment of DNA to be introduced is usually known with the free DNA method, the site where the DNA is integrated cannot be predicted. While the sequence of the starting DNA can be determined with precision, free DNA delivery frequently leads to integration of multiple copies or portions of the gene of interest. Selectable markers, i.e., genes whose expression can be detected soon after the cells have been treated with DNA, are used to recover the very small fraction of cells that are transformed. For example, if the markers confer resistance to a toxic agent, such as an antibiotic or a herbicide added to the culture medium, then only those cells which carry and express the non-host DNA are able to grow.
Another method, more widely used today, employs the bacterial plant pathogen Agrobacterium tumefaciens. In nature, this bacterium infects wounds in broad-leafed plants and induces the formation of tumors or galls. The mechanism of tumor induction using the Agrobacterium method involves the movement of a part of the DNA of a large plasmid carried by the bacterium into some of the host cells. In some of the cells, a host cell chromosome takes up a part of the plasmid DNA, whereupon the plasmid DNA directs the cell to undergo repeated divisions that result in tumor formation. This integrated tumor-inducing DNA also directs the synthesis of an uncommon group of amino acid derivatives (opines) that only the bacterium can use as a source of carbon and nitrogen for further growth. The tumor-inducing DNA can be made nonpathogenic by removing the elements responsible for releasing the controls of cell division and for opine formation. The nonpathogenic DNA (T-DNA), which no longer induces tumor formation, can then be used to carry a different organism’s gene into a host-cell chromosome. As with the free DNA method, cells carrying T-DNA can be detected by incorporating selectable markers such as antibiotic or herbicide resistance. In this way, only cells carrying the resistance markers can grow on culture media in which the antibiotic or herbicide is incorporated; all untransformed cells are killed.
The use of A. tumefaciens greatly increases the precision of DNA insertion. Agrobacterium uses specific DNA-signaling sequences (T-DNA borders) to determine the start and stop points of DNA transfer to plant cells. Although there can still be substantial variation in the transferred DNA, the endpoints of DNA transfer are usually localized to a small region, within 10–100 bases. Moreover, the number of copies of inserted genes can usually be limited to one or a few. Recent improvements in transformation procedures have permitted researchers to largely switch from the free DNA techniques to Agrobacterium. In any case, the precision of rDNA biotechnology permits accurate determination of the location and number of copies of the inserted DNA, even if the location of DNA insertion cannot be controlled.
Scientific knowledge of the structure of the plant genome has grown as a result of research on the “laboratory plant” Arabidopsis thaliana, a small plant in the cabbage family that has only five chromosomes and grows from seed to seed in about seven weeks. Sequencing the entire genome of this plant is now almost complete. Because of the great similarities among plants in general, Arabidopsis can be used as a crop plant analog, and DNA sequences from Arabidopsis of known function can be used to identify their homologs in economic crops. DNA markers can be used to identify chromosome regions that carry blocks of genes of individually small effect, quantitative trait loci or QTLs, which contribute to characteristics such as yield, maturity, baking quality, flavor, and aroma, making possible much more sophisticated selection procedures for plant breeding (McCouch, 1998).
The opportunity to select and multiply a gene of interest and then introduce it into a crop plant was of great interest to most plant breeders because it heralded the era of directed genetic change. It was now possible to introduce a new gene into an accepted and adapted variety in a single step. This reduced the long and tedious process of winnowing out the many forms that are inferior to the adapted varieties that are characteristic products of most conventional breeding programs which introduce new characters from wild unadapted material. In practice, rDNA biotechnology-derived forms can be better thought of as new forms of germplasm to be incorporated into breeding programs, thereby extending the range of characteristics available to a breeder. The breeder must still test the results to ensure that the step of introducing the non-host gene, or transgene, causes no other changes that would be detrimental to the farmer, the consumer, or the environment. As discussed in the Safety section of the report, these tests include detailed analyses of the composition of the product harvested from the rDNA biotechnology-derived form.
--- PAGE BREAK ---
The first rDNA biotechnology-derived food plant marketed in the U.S. was the FlavrSavr™ tomato, introduced in 1994. Produced using T-DNA, this tomato carried an antisense gene for the enzyme polygalacturonase (PG), an enzyme formed as the fruit ripens and which is responsible, in large part, for fruit softening. The gene encoding PG was isolated, inverted in the cloning vector (producing an antisense form), and then introduced into cells that also carry the gene in the normal orientation. In the inverted DNA, the mRNA is transcribed from the wrong DNA strand to form an antisense message. As a result, much less of the enzyme is produced. It was expected that the fruits of the tomato would have an extended shelf life, since they would not soften as rapidly as normal fruit. In fact, the FlavrSavr tomato was not a commercial success as a retail product because of uncompetitive agronomic characteristics; however, a processing variety engineered with a related construct proved to be useful to processors, since the ripe fruit has a higher solids content, resulting in economic and quality advantages.
Following the introduction of the rDNA biotechnology-derived tomato in 1994, other rDNA biotechnology-derived crops that contained modified agronomic traits soon followed. These plants included squash that are resistant to some strains of zucchini yellows and watermelon mosaic viruses in 1994, insect-resistant potato and cotton in 1995 and corn in 1996, and herbicide-tolerant soybean and canola in 1996. Although the consumer’s awareness is largely limited to these products, there are many others under development that are expected to appeal more directly to consumers. These include fruits, root and leaf vegetables, and grains with enhanced nutritional and health-promoting properties.
Recombinant DNA Biotechnology-Derived Foods
Recombinant DNA biotechnology-derived foods are part of the continuing sequence of genetic improvement of the food supply. Although it is sometimes portrayed as fundamentally new, the newness of rDNA biotechnology is best considered from a historical perspective.
The plants and animals that modern agriculture produces today to feed the world’s people are the result of more than 10,000 years of genetic modification and refinement. For example, there is the agricultural green revolution, which has contributed to increased human longevity and improved quality of life in developing countries. The green revolution is viewed by many knowledgeable scientists as the latest major achievement in a long quest begun by ancient agriculturists who first cultivated and domesticated wild plants for food and fiber.
Genetic modification of plants began approximately 10,000 years ago when man first used what is referred to as selective breeding. This technique simply involved saving seeds from the most vigorous plants in an environment for replanting at a later time. Over a period of many years, this selection resulted in higher-yielding varieties of the crop. It is this type of selection that, for example, turned the wild precursor of modern maize, teosinte, into an important human food and animal feed crop in America. The same processes in the Near East—the Fertile Crescent—resulted in einkorn and emmer wheat, barley, lentil, pea, chickpea, and bitter vetch (Lev-Yadun et al., 2000). Likewise, the progenitor of the modern tomato bears almost no resemblance to its modern relatives, which are the result of centuries of selection and DNA recombination at the organism level.
Selective breeding relies principally on sexually transmitted genetic diversity in a starting population. By picking the best or most vigorous plants, breeders over time enrich the genetic makeup of a plant for attributes such as higher yields, increased resistance to pests, and greater compatibility with production schemes. It should be noted that this process in itself runs counter to natural selection. Breeding involves selection for optimal growth for human purposes or other characteristics in an agricultural setting and in many cases is inconsistent with nature and the ability of the organism to survive under evolutionary pressure. Therefore, human intervention has involved what can be called a primitive type of genetic engineering from the outset.
An excellent example of breeding versus natural selection can be gleaned from the history of cultivated wheat. The seeds of wild wheat relatives are dispersed by the shattering of brittle seed heads. In the earliest stages of domestication, 10,000 years ago, forms that do not shatter were selected, which enabled gatherers to collect the ripe seeds rather than pick them up from the ground. Such a mutation in nature would prevent seed dispersal and lead to rapid extinction of those plants in the wild.
--- PAGE BREAK ---
As the available unused genetic diversity of the species diminishes, the potential for improvement also decreases. Since crop improvement relies on genetic diversity, i.e., new sources of genes and expression of existing genes, continued improvement has required and will continue to require even greater diversity. This need for diversity led to the next developments in plant breeding when farmers discovered that crosses between certain closely related species would produce fertile offspring. Cross-breeding (also known as interspecies or intergeneric breeding), either fortuitous or intentional, permitted recombination and selection among genes at a whole new level to provide new sources of genetic diversity and desirable traits.
Interspecies or cross-breeding offers two possible outcomes. First, new species that contain all of the genes from multiple parents can be created. Thus, triticale, a fertile wheat-rye hybrid, became a reality. The first wheat-rye hybrid plants, reported in 1876, were completely sterile, but fifteen years later fertile sectors were reported on a spike that resulted from spontaneous chromosome doubling (Gregory, 1987). Second, another alternative involves recombination, where a single genome is maintained in the offspring, but that genome now consists of randomly chosen copies of genes from either of the parent species. This latter type of breeding in a sense is the precursor to modern rDNA biotechnology; however, it is highly imprecise. Large segments of chromosomes containing thousands of individual genes have been introduced from one species into another in this way. This type of technology is employed today by breeders of many crops, including tomato (discussed below), soybean, canola, and cotton, which are all products of extensive genetic modification and selection.
The products of naturally occurring interspecies crosses have been employed for thousands of years, and many of the foods eaten today are derived from such crosses. A good example is cultivated hexaploid wheat, which has three different genomes, each derived from a wild ancestral species. For thousands of years, this technology has relied upon the ability of a genetic cross to produce fertile offspring. Thus, it is considered “natural.” Many interspecific hybrids are infertile; for example, the original wheat-rye hybrids were sterile, and seeds could only be produced after spontaneous chromosome doubling had taken place. Thus, while interspecific crosses opened up a vast new genetic resource to plant breeders, the need for fertile progeny limited the usefulness of this diversity.
Sometimes, a cross of two species can produce a viable embryo, which develops for a period of time, then degenerates and dies. However, by using the technique known as embryo rescue, the embryo can be recovered shortly after fertilization and placed in an in-vitro tissue culture system. In this artificial setting, the embryo can develop into a mature, fertile plant. Tissue culture can thus expand access to genetic diversity by saving crosses that would not survive outside a laboratory.
Some attention has been paid to the use of ionizing radiation and chemicals to induce mutations and expand the range of variation available to breeders, but very few successful new forms of crop plants have been obtained in this way. The same is true of somaclonal variation arising in tissue culture. However, spontaneous mutations have been important in the development of some cultivated plants.
All of these conventional techniques for crop improvement share the disadvantage that they are, by nature, imprecise and unpredictable and only occasionally useful. Spontaneous and induced mutation can lead to one desirable change and many undesirable collateral changes in an organism’s DNA makeup, which must be selected out. Breeders cannot and do not attempt to define in molecular terms the changes that they make within a genome. Rather, they employ standard selection procedures to screen for new plants with novel alterations and incorporate these plants into their breeding programs. In spite of the undefined nature of these changes, many years of experience have affirmed the safety and usefulness of genetically improved varieties. Plant breeders, farmers, food manufacturers, and consumers all have routine, frequent, and extensive exposure to these genetically improved varieties.
An excellent example of how breeders use all of the above techniques is the tomato. The tomato, Lycopersicon esculentum var. cerasiforme, originates from central Mexico. The original species bears little resemblance to current varieties, which are the result of much genetic manipulation. The growth habits of the plant, resistance to viruses, diseases, and nematodes, as well as fruit taste and appearance are a consequence of mutation, hybridization, and selection. For example, resistances to several diseases, tobacco mosaic virus, and nematodes were introduced from the distantly related species, Lycopersicon peruvianum and Lycopersicon chilense. Crosses between these two species and L. esculentum required embryo rescue. Each new resistance represents the introduction of a large chromosome segment from the distant relative into L. esculentum. The typical introduced non-host DNA segment contains between 100 and 1,000 genes.
--- PAGE BREAK ---
A specific example illustrates the imprecision of traditional breeding. Introduction of resistance to the fungal disease Fusarium crown rot involved a cross between an irradiated L. esculentum variety and L. peruvianum (Rowe and Farley, 1981). From this cross, a resistant plant was selected and used in subsequent breeding. This resistance gene, along with its complement of other genes, is present in many commercial varieties of tomato today. As the tomato is a member of the nightshade family and many of its wild relatives contain high levels of toxicants in the interspecific crosses with L. esculentum, breeders have selected for varieties with minimal toxicant content. While there is no requirement for toxicant screening in traditional tomato breeding programs, it is widely practiced. Moreover, toxicant screening is an integral part of assessing the safety of new rDNA biotechnology-derived varieties.
It is against this experience base that rDNA biotechnology must be examined and compared. Recombinant DNA techniques involve the introduction of one or a few defined genes into a plant. While these introduced genes are often from other, non-host sources, the introduction of non-host DNA is not novel. In fact, remnants of an ancient Agrobacterium transformation have been identified in Nicotiana species (Furner et al, 1986). It is important to note that it is the very same Agrobacterium that is now used widely by researchers to introduce genes into plants.
Similarly, microorganisms have been used in food technology for thousands of years. As early as 6000 B.C., Sumerians and Babylonians used yeast to brew beer. Although the ancients knew nothing about microorganisms and could not knowingly culture them, they nevertheless systematically selected those with desirable fermentation characteristics to improve their food. In modern times, the increasingly powerful science of genetics has been systematically applied to produce many valuable variants of yeast and bacteria.
Recombinant DNA techniques have provided both an important new set of tools and access to a broader range of markets. They enable researchers seeking specific plant characteristics to precisely identify, characterize, enhance, and transfer the appropriate individual genes rather than uncontrolled and randomly assorted groups of genes, hoping the desired ones were included. Researchers can now readily move selected and well-characterized genetic material from virtually any source in nature, greatly increasing the diversity of useful genes available for crop and microbe improvement. The long, continuous search for improved plants and the benefits of useful microorganisms is now increasingly based on the use of rDNA biotechnology techniques.
Microorganisms are used in the production of foods, beverages, industrial detergents, antibiotics, organic solvents, vitamins, amino acids, polysaccharides, steroids, and vaccines. Practical applications of pre-rDNA biotechnology include a variety of organisms used in pest control (including many that are themselves often considered to be pests, in other settings, e.g., preparations of the bacterium Bacillus thuringiensis sold at most garden supply stores). Biological agents are also used as growth promoters for plants. Preparations containing the bacterium Rhizobium, which fixes atmospheric nitrogen, converting it into nitrogen-containing ions that are essential plant nutrients, have been sold in the U.S. since the late 19th century. As early as the mid-1980s, these pre-rDNA biotechnology products, together, had a value in excess of $100 billion annually (Anonymous, 1985). Since the introduction of rDNA biotechnology, many of these microorganisms have been improved, such as those used to produce the enzyme chymosin necessary for cheese production.
Some critics of rDNA biotechnology have taken the view that it represents a fundamental change from traditional techniques for the genetic modification of plants and microorganisms. In a 1989 report, the National Research Council considered and rejected this argument:
However, no conceptual distinction exists between genetic modification of plants and microorganisms by classical methods or by molecular techniques that modify DNA and transfer genes. . . . The same physical and biological laws govern the response of organisms modified by modern molecular and cellular methods and those produced by classical methods.
The NRC went on to characterize rDNA biotechnology as part of a sequence of scientific advances that has extended over a 10,000-year period (NRC, 1989).
--- PAGE BREAK ---
A 1991 joint Food and Agriculture Organization/World Health Organization consultation, addressing the question of the safety of rDNA biotechnology-derived foods, came to similar conclusions (FAO/WHO, 1991):
Biotechnology has a long history of use in food production and processing. It represents a continuum embracing both traditional breeding techniques and the latest techniques based on molecular biology. The newer biotechnological techniques, in particular, open up very great possibilities of rapidly improving the quantity and quality of food available. The use of these techniques does not result in food which is inherently less safe than that produced by conventional ones.
A timeline that shows the increasing power of genetic modification over the past 12,000 years appears in Fig. 1. 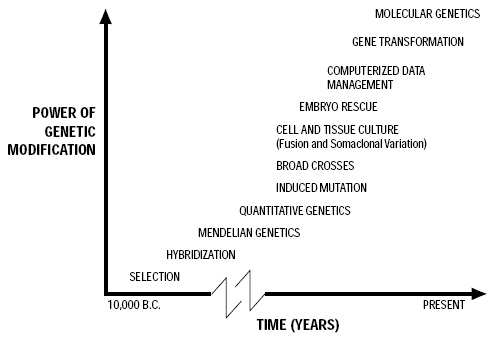
Even though food derived from biotechnology in the broad sense is hardly new, some critics nevertheless have been concerned that rDNA biotechnology may result in different and dangerous organisms. Considering that there are tens of thousands of the host organism’s own genes, the introduction by precise techniques of one or a few additional, well-characterized genes does not create an organism that is more likely to be changed in gross physical properties or wholesomeness than an organism derived through a traditional breeding program. Indeed, because of the greater precision in selecting the desired trait, an adverse result is unlikely. A corn plant with a newly inserted bacterial gene that confers increased resistance to the European corn borer (a commercially important insect predator) is still a corn plant. Likewise, a microorganism long used for food production is not altered in any fundamental way by the insertion of additional copies of a gene-encoded rate-limiting enzyme. Aided by the recent voluminous data from the DNA sequencing of various genomes and other basic research on plants, such questions have been widely discussed and reported by an array of national and international scientific groups. Their conclusions are discussed in the Safety section of the report.
Consider whether genetic recombination, itself, is of concern. It has already been established that people have long engaged in the systematic improvement of domesticated microbes, plants, and animals. But the impact and importance of these changes are much smaller than what occurs continuously in nature. Innumerable recombinations between related and unrelated organisms have occurred by several mechanisms. Sexual reproduction randomly combines genes from two parents in the offspring, which then has a unique set of genes to pass along to the next generation. In the gut, decomposing tissue, and infected wounds, bacteria take up naked mammalian DNA, albeit inefficiently, when they encounter disintegrating cells, and some of this DNA may be incorporated into the bacterial genome, but there is no established evidence that this happens (Davis, 1986). Over the past million years and longer, mammalian-bacterial genetic hybrids have appeared, been tested by competition within bacterial populations and by environmental stresses, and conserved or discarded by natural selection. Similar genetic recombination and hybridization also has been widespread among fungi, viruses, and plants.
Evolutionary biology provides data relevant to the issue of the uniqueness of chimeric genes (genes containing modified or substituted control signals joined to portions of the native genetic information) created by rDNA biotechnology. Does the transfer into a squash of a viral gene to confer viral resistance affect its “squashness” or transfer “viralness” to the new hybrid? The sequencing of various genomes during the past decade has revealed that nature has been remarkably conservative about maintaining and using effective molecules as they evolved. Similar protein sequences and biochemical pathways are found in different species, across genera, and even across phylogenetic kingdoms. The Escherichia coli genome, for example, contains gene sequences that are closely related to those in a wide spectrum of organisms, ranging from other bacteria to plants, insects, amphibia, birds, and humans.
Another issue, conversion of a non-pathogen into a pathogen through limited genetic recombination, is best considered within the context of the nature of pathogenicity. This process is both complex and multifactorial. Pathogenicity usually is not a trait produced by a single gene; however, the transfer of a single gene to an organism that has all the other necessary genes can make it pathogenic. Pathogenicity requires the coordinated activity of a set of genes that affect essential properties.
--- PAGE BREAK ---
A pathogen must possess three general characteristics, each of which involves multiple genes. First, pathogens must survive and be able to multiply or produce toxin in or upon host tissues or food sources. This necessitates an appropriate oxygen tension, pH, temperature, water activity, and nutritional milieu. Pathogens must be able to adhere to specific surfaces on or in the host. Second, the pathogen must be able to resist or avoid the host’s defense mechanisms for the period of time necessary to multiply to sufficient levels to cause disease. Third, the pathogen must be able to survive outside of the host and must be disseminated to new host organisms. The organism must be meticulously adapted to this pathogenic lifestyle. On the other hand, a mutation that interferes with a gene essential to any one of the three characteristics of a pathogen can eliminate pathogenicity. It is worth noting that severe pathogenicity is even more dependent upon favorable conditions and is, therefore, much rarer in nature than mild pathogenicity.
The probability of creating and commercializing an organism inadvertently capable of producing a medical or agricultural problem is therefore quite small. The expert panels are of the view that this probability is lower with rDNA biotechnology than with the more random, less targeted, and less predictable traditional methods of genetic modification. In rDNA biotechnology-derived organisms, typically one, two, or three genes are being inserted. The genes, gene products, and their functions are known. This information guides scientists in determining which possible risks are relevant and need to be explored. In comparison, with traditional breeding, a large number of genes with unknown functions are involved, making it much more difficult to sort through the progeny and focus on the relevant risks involved.
Adverse outcomes accompanying genetic change have always been possible but are routinely intercepted during the usual, extensive testing that takes place in growth chambers, greenhouses, and the field. Whatever the technique used to craft a variety, it goes through extensive testing before being used commercially, particularly if the developer chooses to enter it into formal seed registration programs. In practice, the testing is even more extensive in the case of an rDNA biotechnology-derived variety. Therefore, the expert panels are of the view that rDNA biotechnology has the potential to reduce still further the chance that any such mishap will occur. The field and chemical testing that accompany it—even more thorough than in traditional genetic modification—make such an unfavorable outcome even more unlikely. As noted earlier, genetic changes that make a plant more useful to humans usually have made the plant less “fit” and less able to survive in the wild.
Federal Regulation of rDNA Biotechnology
Regulatory oversight over rDNA biotechnology spans three major federal agencies: the Food and Drug Administration (FDA), the Environmental Protection Agency (EPA), and the U.S. Department of Agriculture (USDA). Jurisdiction over the varied rDNA biotechnology products is determined by their use, as has been the case for products made by traditional means. More than one agency may be involved in regulating different aspects of an rDNA biotechnology-derived product. As the regulatory mandate varies, so does the nature of the agencies’ risk assessment and management protocols.
The “Coordinated Framework for Regulation of Biotechnology,” prepared by the White House’s Office of Science and Technology Policy (OSTP) and published in the Federal Register of June 26, 1986 (51 FR 23302), is the current comprehensive federal policy for ensuring the safety of rDNA biotechnology research and products. It established the principles and procedures for coordination and jurisdiction among federal agencies for the oversight of rDNA biotechnology. Subsequently, the OSTP prepared and published in the Federal Register of February 24, 1992 (57 FR 6753) “Exercise of Federal Oversight within Scope of Statutory Authority: Planned Introductions of Biotechnology Products into the Environment.” This notice described a risk-based, scientific approach to the oversight of planned introductions of rDNA biotechnology-derived products into the environment, focusing on the characteristics of the product and the environment into which it is being introduced, not the process by which the product is created.
The ultimate goal of the OSTP policy is to ensure the overall safety to humans and the environment of, in relevant part, foods, food ingredients, and feeds produced using rDNA biotechnology. In an April 2000 report, the National Research Council stated: “In general, the current U.S. coordinated framework has been operating effectively for over a decade” (NRC, 2000).
--- PAGE BREAK ---
Although the approach outlined in the 1986 and 1992 OSTP regulatory policy guidelines states that federal policies should be risk-based—i.e., should focus on the risk-related characteristics of products, rather than on the process used—that principle has not been followed by regulatory agencies. The fundamental approach by the federal government to the review and regulation of rDNA biotechnology-derived products has largely been through a process-based trigger to oversight. As discussed below, crops and microbes produced using rDNA biotechnology have been consistently subjected to higher requirements and standards than those applied to similar products produced using traditional techniques (Miller, 1997, 2000). At this time, there is less experience with rDNA biotechnology-derived products, but that experience base is increasing substantially.
Food and Drug Administration
FDA regulates different aspects of rDNA biotechnology under the authority of the Federal Food, Drug, and Cosmetic Act (FFDCA) and the Public Health Service Act (PHSA). FDA has a mandate to ensure the safety of all food (except for meat and poultry products) sold in the U.S., as well as the safety and efficacy of pharmaceutical products. To date, FDA has conducted almost fifty reviews of rDNA biotechnology-derived plant products used for human food or animal feed.
• Human Food and Animal Feed. Except for meat and poultry products regulated by USDA, FDA is responsible for ensuring the safety and proper labeling of food products for human consumption. FDA also regulates the safety and labeling of animal feed, taking into account both the safety to human consumers of animal-derived food products and the safety to the animal being fed. FDA’s statutory authority is provided by the FFDCA. FDA’s framework for the regulation of food labeling is discussed in the Labeling section of the report; the framework for the regulation of food safety is discussed below.
FDA has very broad authority to regulate the introduction of new food crops, whether conventionally grown, produced through hybridization or cross-breeding, or produced using rDNA biotechnology. Every firm or individual that produces whole foods or food ingredients is legally required to ensure the safety of foods and food ingredients introduced into commerce. FDA has a number of enforcement tools that can be used to ensure the safety of food. Specifically, the FFDCA prohibits the adulteration of any food item that moves in interstate commerce (21 USC §342). Of particular importance, foods are deemed adulterated if they contain certain poisonous and deleterious substances (21 USC §342(a)(1)). With certain exceptions that are not relevant to this discussion, the FFDCA defines a “food additive” as any substance, not “generally recognized as safe” (GRAS) by qualified experts for its intended use, that becomes a component or otherwise affects the characteristics of food (21 USC §321(s)). Food additives must be the subject of a petition to FDA, followed by FDA premarket approval; their manufacturers have the burden of establishing, through scientific testing, the safety of the substances (21 USC §348). In comparison, a food manufacturer that believes its food ingredient is GRAS may market the ingredient without seeking FDA’s concurrence, subject to the risk that FDA will disagree and take legal action to remove the ingredient from the marketplace.
In the U.S., whole foods such as fruits, vegetables, and grains are not regulated as “food additives” and are not required to undergo premarket approval; nor are they commonly subjected to extensive safety testing. Thus, new varieties of crop plants produced by traditional breeding methods are not subject to FDA premarket review. Nevertheless, authority exists to ensure that such foods do not present a reasonable possibility that consumers might be injured by consuming them. With respect to all foods, FDA can initiate legal action to remove a food from the market if it is judged to present a health risk. While there is no evidence that such authority has ever needed to be exercised with respect to traditional breeding practices, plant breeders and food processors have several times intercepted toxic food plants before they reached the market. An example, mentioned in the Safety section of the report, is the Lenape potato.
On May 29, 1992, FDA published a policy statement (57 FR 22983) on foods and animal feed derived from new plant varieties developed by conventional and new breeding techniques, including rDNA biotechnology techniques. FDA stated:
--- PAGE BREAK ---
This policy statement is a clarification of FDA’s interpretation of the Federal Food, Drug, and Cosmetic Act (the act) with respect to technologies to produce foods, and reflects FDA’s current judgement based on new plant varieties now under development in agricultural research. This action is being taken to ensure that relevant scientific, safety, and regulatory issues are resolved prior to the introduction of such products into the marketplace.
FDA set forth its authority to control food products derived by rDNA biotechnology techniques and listed the safety issues that need to be addressed in assessing the safety of whole foods that contain or use rDNA biotechnology-derived plants and microorganisms. One key point is that under certain conditions, foods and food ingredients derived from rDNA biotechnology-derived plants or microorganisms may be subject to the provisions of existing requirements governing food additives and GRAS substances. FDA noted that in the case of foods derived from new plant varieties, it is the transferred genetic material and intended expression product(s) that could be subject to food additive requirements if these materials are not GRAS. FDA stated that if the intended expression product is a protein, carbohydrate, or other substance that differs substantially from substances currently present in food, then that substance might not be GRAS and may be a food additive requiring premarket approval. Another important point is that if an rDNA biotechnology-derived plant or microorganism is used to produce a GRAS substance or an approved food additive, the resulting material would continue to be regulated in a similar fashion to the way in which it has historically been regulated.
FDA’s 1992 policy on new plant varieties applies irrespective of whether the plant arose from rDNA biotechnology or “conventional” genetic modification methods. FDA does not routinely subject foods from new plant varieties to a premarket approval process or to extensive scientific safety tests. FDA’s policy does, however, define certain safety-related characteristics of new foods—such as transfer of an allergen or increased levels of a natural toxicant—that trigger additional scrutiny. FDA’s policy includes a flow chart (Fig. 2) for guidance that asks a series of questions directed to scientific issues of safety and nutrition of the foods derived from the new plant variety. The assessment focuses on the following risk-based considerations:
- Toxicants known to be characteristic of the host and donor species.
- The potential that food allergens will be transferred from one food source to another.
- The concentration and bioavailability of important nutrients for which a food crop is ordinarily consumed.
- The safety and nutritional value of newly introduced proteins.
- The identity, composition, and nutritional value of modified carbohydrates, fats, or oils.
Fundamentally, FDA’s current (1992) policy is that existing requirements mandate the same safety standards for foods, food ingredients, and feeds, regardless of the techniques used in their production and manufacture. Nevertheless, FDA has maintained a “voluntary consultation procedure,” in which producers of rDNA biotechnology-derived foods are asked to consult with the agency before marketing their products, and without exception they have done so (HHS, 2000). To date, almost 50 new rDNA biotechnology-derived foods have been evaluated successfully in FDA’s voluntary consultation process. These evaluations are summarized in Table 1. Each entry represents a separate consultation, and each consultation may represent more than one line of the traits indicated. Products are grouped by the year in which their consultations were completed. The trait introduced into the variety plus the origin and identity of the introduced gene responsible for the trait are given (FDA, 2000).
FDA’s official policy may change significantly, as the Clinton Administration announced in May 2000 that FDA will publish a proposed rule that would require producers to notify FDA 120 days before marketing an rDNA biotechnology-derived food and provide the agency with data that affirm the new food’s safety. In practice, assuming that new regulatory requirements are proposed and finalized, FDA’s current voluntary consultation procedure would become mandatory.
--- PAGE BREAK ---
• Pharmaceuticals and Human Vaccines. FDA regulates rDNA biotechnology-derived pharmaceutical products for human and animal use under the FFDCA and the PHSA. FDA also regulates rDNA biotechnology-derived vaccines for human use under the PHSA, while USDA regulates vaccines for animal use. Under both the FFDCA and the PHSA, new products must be the subject of premarket approval, based on laboratory and clinical testing to show the safety and effectiveness of the products for their intended uses (21 USC §§355 and 360b; 42 USC §262). 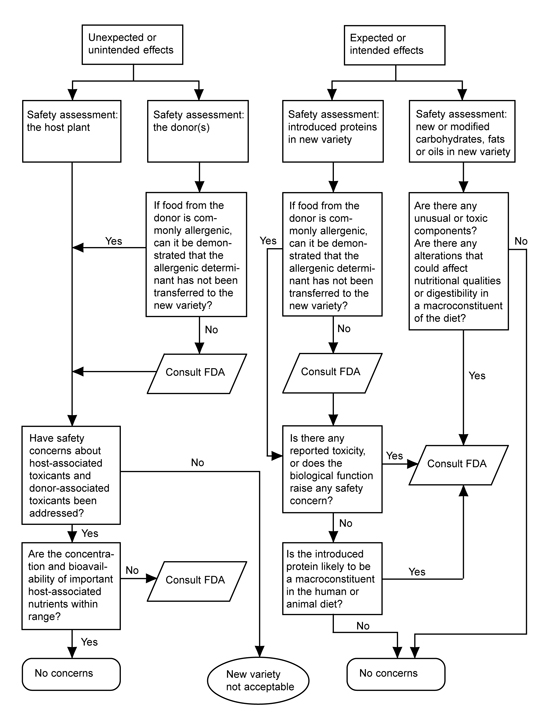
U.S. Department of Agriculture
Two USDA agencies are relevant to the regulation of foods and other products derived using rDNA biotechnology.
• Foods. The Food Safety and Inspection Service (FSIS) is responsible for regulating the safety and labeling of meat and poultry products for human consumption. FSIS consults with FDA regarding the safety of food ingredients. Because transgenic animals are beyond the scope of this report, USDA’s regulation of meat and poultry products will not be discussed further.
The Animal and Plant Health Inspection Service (APHIS) is the agency within the USDA charged with protecting American agriculture against pests and diseases. Under the Plant Quarantine Act (PQA, 7 USC §151) and the Federal Plant Pest Act (FPPA, 7 USC §150), APHIS can regulate the importation and interstate movement of plants and plant products that may result in the entry into the U.S. of injurious plant diseases or insect pests.
The field-testing and the commercial sale of agricultural rDNA biotechnology-derived crops are regulated by APHIS through a permit and notification system. USDA’s regulations (7 CFR Part 340) cover the introduction of organisms and products altered or produced through genetic engineering which are plant pests or for which there is reason to believe are plant pests. “Plant pests” include agents that can directly or indirectly injure or cause disease or damage in or to any plant. A “regulated article” includes any organism or any product, which has been altered or produced through rDNA biotechnology, which is a plant pest, or for which there is reason to believe is a plant pest. The permit and notification system does not apply to plants that are modified through traditional breeding methods. Thus, USDA’s regulatory protocol is process based.
The introduction of a regulated article is prohibited unless a permit under 7 CFR Part 340 authorizes the introduction. The regulation is intended to prevent the introduction, dissemination and establishment of plant pests in the U.S. APHIS will grant a permit only if it determines that the plant poses no significant risk to other plants in the environment and is as safe to use as more traditional varieties. APHIS can authorize nonregulated status for an article through a petition for a “determination of nonregulated status.” Nonregulated status allows a plant to be treated like any other plant, i.e., allows for the plant to be widely grown and commercialized.
• Animal Vaccines. APHIS regulates animal vaccines under the Virus-Serum-Toxin Act (21 USC §§151–159). In general, animal vaccines are subject to premarket approval, based on testing to show their safety and effectiveness.
Environmental Protection Agency
EPA’s stated mission is to protect human health and to safeguard the natural environment—air, water, and land—upon which life depends. EPA’s responsibilities under the Federal Insecticide, Fungicide, and Rodenticide Act (FIFRA, 7 USC §§136–136r) for registering pesticides, setting environmental tolerances for pesticides, and establishing exemptions for pesticide residues in and on crops are relevant to rDNA biotechnology-derived foods. A pesticide is any substance or mixture of substances intended for preventing, destroying, repelling, or mitigating any pest.
The Food Quality Protection Act (FQPA) of 1996 amended FIFRA and the FFDCA by establishing a single, health-based standard for assessing the risks of pesticide residues in food or feed. The standard measures the aggregate risk from dietary exposure and other non-occupational sources of exposure. EPA must now focus explicitly on exposures and risks to infants and children, assuming when appropriate, an additional safety factor to account for uncertainty in data.
--- PAGE BREAK ---
If EPA determines that there is a “reasonable certainty that no harm” to the public will result from aggregate exposure to a particular pesticide residue, then that residue level will be deemed “safe.” 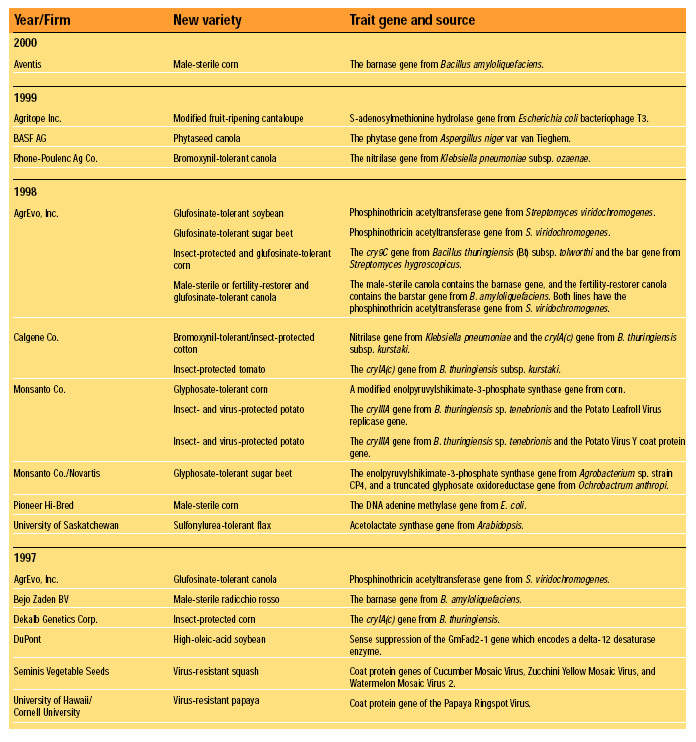
 In the case of pesticides produced by plants developed using rDNA biotechnology, EPA’s November 23, 1994 (59 FR 60519), proposed rule takes the view that its regulatory process is focused on the pesticide and not on the plant; plants are subject to regulation only if they produce plant pesticidal proteins as a result of modification with rDNA techniques. Although EPA has not finalized that proposed rule, EPA has been implementing its essential elements since 1995 (NRC, 2000). EPA’s evaluation of products of rDNA biotechnology is distinct from the procedures used to assess the safety of the products of more conventional technology. In April 2000, the NRC issued a report after evaluating the science and regulation of rDNA biotechnology-derived pest-protected plants. The NRC panel accepted without critical evaluation the EPA’s regulatory approach. In contrast, eleven major scientific societies representing more than 80,000 biologists and food professionals published a report warning that the EPA policy would discourage the development of new pest-resistant crops and prolong and increase the use of synthetic chemical pesticides; increase the regulatory burden for developers of pest-resistant crops; limit the use of biotechnology to larger developers who can pay the inflated regulatory costs; and handicap the U.S. in competition for international markets.
In the case of pesticides produced by plants developed using rDNA biotechnology, EPA’s November 23, 1994 (59 FR 60519), proposed rule takes the view that its regulatory process is focused on the pesticide and not on the plant; plants are subject to regulation only if they produce plant pesticidal proteins as a result of modification with rDNA techniques. Although EPA has not finalized that proposed rule, EPA has been implementing its essential elements since 1995 (NRC, 2000). EPA’s evaluation of products of rDNA biotechnology is distinct from the procedures used to assess the safety of the products of more conventional technology. In April 2000, the NRC issued a report after evaluating the science and regulation of rDNA biotechnology-derived pest-protected plants. The NRC panel accepted without critical evaluation the EPA’s regulatory approach. In contrast, eleven major scientific societies representing more than 80,000 biologists and food professionals published a report warning that the EPA policy would discourage the development of new pest-resistant crops and prolong and increase the use of synthetic chemical pesticides; increase the regulatory burden for developers of pest-resistant crops; limit the use of biotechnology to larger developers who can pay the inflated regulatory costs; and handicap the U.S. in competition for international markets.
Summary
In this section, the general concept of biotechnology has been introduced and the scope of the overall report has been defined. Further, extensive background information has been provided to assist the reader in understanding rDNA biotechnology-derived foods. Biotechnology has been discussed in considerable detail, and the point has been made that, in the view of many knowledgeable scientists, rDNA biotechnology-derived foods are the latest major step in a 10,000-year process of genetic improvement of food. Finally, this section has discussed federal regulation and oversight of rDNA biotechnology.
This section has provided the foundation for the three sections that follow. The sections are based on a review of the scientific literature on three different but related aspects of rDNA biotechnology-derived foods—human food safety, benefits and concerns, and labeling—and the public policy implications of the underlying science. In developing this state-of-the-science report, it is IFT’s intent to promote a meaningful public discussion of the subject that is based on sound science.
Key documents referenced in the report and other biotechnology resources
Food and Drug Administration (FDA) Center for Food Safety and Applied Nutrition
• Biotechnology main page: vm.cfsan.fda.gov/~lrd/biotechm.html
• 1992 policy statement: vm.cfsan.fda.gov/~acrobat/fr920529.pdf
• Guidance on current consultation procedures: vm.cfsan.fda.gov/~lrd/consulpr.html
U.S. Department of Agriculture (USDA)
• Agency regulation of biotechnology: www.aphis.usda.gov/biotechnology/index.html
• Biotechnology resources from the National Agricultural Library (NAL): www.nal.usda.gov/bic
• NAL Internet resources and links: www.nal.usda.gov/bic/www.html
National Research Council (NRC)
• 2000 report on genetically modified pest-protected plants: books.nap.edu/catalog/9795.html
• 2000 report on transgenic plants and world agriculture: bob.nap.edu/html/transgenic/notice.html
• 1989 report on field testing of GMOs: www.nap.edu/books/0309040760/html
Food and Agriculture Organization of the United Nations (FAO)
• Statement on biotechnology: www.fao.org/biotech/state.htm
• Biotechnology resources: www.fao.org/waicent/faoinfo/agricult/guides/subject/b.htm
• 1996 joint FAO/WHO consultation, Biotechnology and Food Safety: www.fao.org/waicent/faoinfo/economic/esn/biotech/tabconts.htm
World Health Organization (WHO)
• Genetically modified food main page, including information about Codex Alimentarius activities: www.who.int/fsf/gmfood/index.htm
• 2000 joint FAO/WHO consultation, Safety Aspects of Genetically Modified Foods of Plant Origin: www.who.int/fsf/gmfood/fao-who_consultation_report_2000.pdf
• 1990 FAO/WHO joint consultation, Strategies for Assessing the Safety of Foods Produced by Biotechnology: www.who.int/faf/gmfood/bio1991repo.pdf
Organization for Economic Co-operation and Development (OECD)
• Biotechnology and food safety main page: www.oecd.org/subject/biotech
• 1993 report on safety evaluation of biotech foods: www.oecd.org/dsti/sti/s_t/biotech/prod/modern.htm
• Biotechnology publications main page: www.oecd.org//ehs/icgb/biopubs.htm
Institute of Food Technologists (IFT)
• Main page: www.ift.org
• Backgrounder on Genetically Modified Organisms: www.ift.org/resource/pdf_files/gmoback.pdf
American Dietetic Association (ADA)
• Position statement on food biotechnology: www.eatright.org/abiotechnology.htm
Council for Agricultural Science and Technology (CAST)
• Biotechnology communications: www.cast-science.org/biotechnology/index.html
International Food Information Council (IFIC)
• Main page: www.ificinfo.health.org
References
Anonymous. 1985. Health impact of biotechnology: Report of a WHO Working Group. Swiss Biotechnol. 2: 7-16.
Avery, O.T., MacLeod, C.M., and McCarty, M. 1944. Studies on the chemical nature of the substance inducing transformation of Pneumococcal types. J.Exp.Med. 79: 137-158.
Davis, B.D. 1986. Evolution, epidemiology, and recombinant DNA. In “Storm Over Biology,” pp. 271-273. Prometheus Books, Buffalo.
FAO/WHO. 1991. Strategies for assessing the safety of foods produced by biotechnology. Report of a Joint FAO/WHO Consultation. Food and Agriculture Org./World Health Org.. World Health Org., Geneva.
FDA. 2000. “Foods Derived from New Plant Varieties Derived through Recombinant DNA Technology.” Food and Drug Administration, Center for Food Safety and Applied Nutrition, Washington, D.C. (http://vm.cfsan.fda.gov/~lrd/biocon.html).
Furner, I., Huffman, G., Amasino, R., Garfinkel, D., Gordon, M., and Nester, E. 1986. An Agrobacterium transformation in the evolution of the genus Nicotiana. Nature 319: 422-427.
Grace, E.S. 1997. “Biotechnology Unzipped: Promises and Realities.” Joseph Henry Press, Washington, DC.
Gregory, R.S. 1987. Triticale breeding. In “Wheat Breeding: Its Scientific Basis,” ed. F.G.H. Lupton, pp. 269- 286. Chapman & Hall, London.
HHS. 2000. FDA to strengthen pre-market review of bioengineered foods. Press release, U.S. Dept. of Health and Human Services, Washington, D.C., May 3.
Lander, E.S. and Weinberg, R.A. 2000. Genomics: Journey to the center of biology. Science, March 10, p. 287.
Lev-Yadun, S., Gopher, A., and Abbo, S. 2000. The cradle of agriculture. Science 288: 1602-1603.
Linn, S. and Arber, W. 1968. Host specificity of DNA produced by Escherichia coli. X. In vitro restriction of phage Fd replicative form. Proc. Natl. Acad. Sci. 59: 1300-1306.
McCouch, S. 1998. Toward a plant genomics initiative: Thoughts on the value of cross-species and crossgenera comparisons in the grasses. Proc. Natl. Acad. of Sciences. 95: 1983-85.
Miller, H.I. 1997. Chpt. 3 in “Policy Controversy in Biotechnology: an Insider’s View.” R.G. Landes Co. and Academic Press, Austin, Tex.
Miller, H.I. 2000. Anti-biotech sentiment has its own risks. Financial Times, March 22, p. 10.
NRC. 1989. “Field Testing Genetically Modified Organisms: Framework for Decisions.” Natl. Res. Council. National Academy Press, Washington, D.C.
NRC. 2000. “Genetically Modified Pest-Protected Plants: Science and Regulation.” Natl. Res. Council. National Academy Press, Washington, D.C.
OTA. 1984. Commercial biotechnology: An international analysis. OTA-BA-218, p. 3. U.S. Congress, Office of Technology Assessment. U.S. Govt. Printing Office, Washington, D.C.
Rowe, R.C. and Farley, J.D. 1981. Strategies for controlling Fusarium crown and root rot in greenhouse tomatoes. Plant Disease Repts. 65: 107-108.
Watson, J.D. and Crick, F.H.C. 1953. Molecular structure of nucleic acid. A structure for deoxyribose nucleic acid. Nature 171: 737-738.
Watson, J.D. and Tooze, J. 1981. “The DNA Story: A Documentary History of Gene Cloning.” W.H.Freeman, San Francisco.
