DNA Test Identifies Animal Species in Food Products
Identifying what animal species a food product contains is important for health, economic, religious, and legal reasons. A microarray test detecting DNA sequences allows species composition—and authenticity or adulteration—to be determined.
It’s not always easy to identify a contaminating species in food. Animal meats often undergo a tremendous amount of processing before being sold as the final food product on supermarket shelves. The end product rarely resembles its whole animal predecessor, making it extremely difficult to distinguish between species that might be present, often allowing contamination to go undetected and food authenticity to go unchecked.
 The presence of unwanted or unknown animal species in food can have a range of effects from benign to deathly serious and is of great concern for public health, economic, religious, and legal reasons. For example, identifying species composition is important to prevent allergic reactions to certain animal products. Unknowingly ingesting a particular allergen can be a serious health danger. Additionally, the unknown presence of any animal ingredients in a vegetarian food may cause individuals to unknowingly violate their belief system. And substitution of an inferior animal species in a food product, often referred to as “food fraud,” has both economic and legal ramifications.
The presence of unwanted or unknown animal species in food can have a range of effects from benign to deathly serious and is of great concern for public health, economic, religious, and legal reasons. For example, identifying species composition is important to prevent allergic reactions to certain animal products. Unknowingly ingesting a particular allergen can be a serious health danger. Additionally, the unknown presence of any animal ingredients in a vegetarian food may cause individuals to unknowingly violate their belief system. And substitution of an inferior animal species in a food product, often referred to as “food fraud,” has both economic and legal ramifications.
Manufacturers and consumers alike have until recently been unable to examine the composition of food at a molecular level. However, for the first time, bioMérieux, a world leader in in-vitro diagnostics located in Marcy l’Etoile, France, has applied microarray technology to develop the FoodExpert-ID® system, launched in late February of this year. Powered by GeneChip® technology developed by Affymetrix, Inc., the microarray (Fig. 1) enables scientists to simultaneously identify the absence or presence of 33 different species of animals in virtually any sample. The array identifies each organism by detecting DNA sequences specific to each animal, allowing species composition to be determined and thereby safeguarding the purity and authenticity of food.
Microarray Analysis of Food
The basic element shared by every member of the food chain is DNA. While many DNA sequences are shared between species, certain gene sequences are different. By measuring these known variable genetic sequences, scientists can distinguish one species’ DNA from anothers’. The microarray is designed to simultaneously and specifically detect different gene sequences, making it a natural technology to distinguish between animal species that might be present in food.
Once exclusively used for basic scientific research, production flexibility and scalability have transformed the microarray into a tool that is being developed for diagnostic purposes as well. The first diagnostic application was envisioned for the clinic, to detect biological abnormalities and improve human health. However, advances in production technology are now allowing for arrays to be produced with sufficient processing power, size, and cost to be used in nonclinical diagnostic settings as well. Food testing represents the first use of microarray technology outside disease and drug-discovery applications.
The incredible potential of high-density microarrays results from a fabrication method that marries semiconductor photolithographic manufacturing technology, combinatorial chemistry, fluorescence spectroscopy, and molecular hybridization. While microarrays currently use millions of DNA probes to measure gene expression or sequence diversity, the fundamental technology allows for the synthesis of more than 1015 distinct probes in the same number of production steps—more than 10,000 times more probes than there are stars in the Milky Way galaxy. And by continuing to refine photolithographic manufacturing, it is possible to include larger subsets of these probes and increase the genetic processing power of each individual array. These advances not only create more powerful arrays, but also make genome discovery routine and affordable. Across multiple research disciplines, whole genome expression analysis and DNA sequence analysis are helping scientists to stratify disease, predict patient outcome, and make better therapeutic choices. Originally a scientific curiosity, the microarray has become an indispensable tool for biological research and offers similar promise for food analysis.
--- PAGE BREAK ---
How the Microarray Measures Food Composition
Microarrays can now be used to safeguard food beyond the existing limitations of established methods, which are unable to detect for the presence of multiple contaminating animal species. For example, X-rays have been used to guarantee food purity against foreign objects such as metal particles, and regulatory agencies have developed exhaustive systems to detect pathogens, such as Escherichia coli and Salmonella, in meat and poultry products. In both Europe and the United States, strict guidelines require that all ingredients present in a food product be defined and identified on the product label. But manufacturers and consumers have been limited by the lack of methods to determine food composition and detect the presence of certain desired animal species or undesired contaminating species.
The basic problem is that when animals are processed into food, the end result often bears little resemblance to the source from which it was rendered. But, while the form and appearance may be unrecognizable, the DNA from the animal remains the same.
The microarray works by detecting species-specific DNA for 33 different animals, including 12 mammals, 5 birds, and 16 fish (Table 1). The system detects the unique DNA sequence for the cytochrome b gene (cytb) present in each different organism. Over the course of evolution, the DNA sequence—the order of adenine, thymine, cytosine, and guanine molecules—of the cytb gene has mutated, creating variations of the same gene in each different animal species. By using the microarray to identify those species-specific gene sequences, scientists can tell which variations of the cytb gene are in a sample, allowing them to determine what animals have been processed in the food under study.
The microarray itself uses more than 80,000 DNA probes, each 17 bases long, to distinguish between multiple cytb gene sequences that may be present in the sample. Once a sequence is determined, it is compared against a known cytb species list and a positive animal identification can be made. Because microarrays are able to simultaneously determine the absence or presence of multiple genes, a single experiment indicates whether each of the 33 species is absent from or present in the original sample.
Microarray Basics
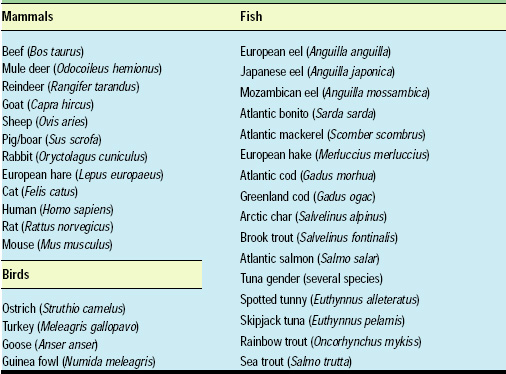
There are only four molecules (“bases”) in every DNA chain: adenine (A), guanine (G), thymine (T) and cytosine (C). These four bases form specific bonds with their molecular counterpart: C partners with G and T partners with A. Pairing is a natural state for DNA and is primarily responsible for its unique double helix shape. If the double helix is pulled apart, it would inevitably move back together, like two long chains of magnets that are attracted to each other.
RNA is also composed of four bases: A, G, C, and uracil (U) instead of thymine. Uracil is related to its DNA equivalent, thymine, such that in RNA, C partners with G and U partners with A. However, unlike DNA, RNA is usually single stranded, meaning that it can easily bind to any other single-stranded matching sequence, whether DNA or RNA.
As indicated in Fig. 2, when a single strand of DNA (ATCATG) matches a strand of RNA (UAGUAC), the two strands are “complementary” and will stick to each other. However, if the bases aren’t complementary, they won’t fit together. An A won’t pair with another A or with a C or a G. Even a single base that doesn’t match its partner could keep a strand from sticking to another single strand.
--- PAGE BREAK ---
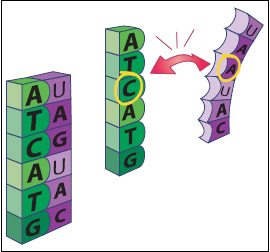
The microarray uses this base pairing—a process known as hybridization—to help researchers identify unknown DNA sequences that might be present in a sample. Because C always pairs with G, and T always pairs with A, if we know that one side of the chain is ATCATG, we don’t need to see the other side to know that it is TAGTAC. If that second strand is RNA, the sequence will be UAGUAC.
Creating the Microarray
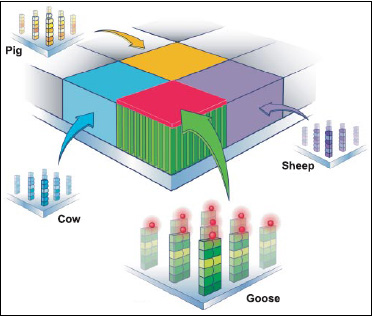
The first step in creating a food-testing microarray is to build a short DNA strand representing a unique animal sequence—i.e., a probe—onto the surface of a glass plate. The short probe sequences are selected to specifically match a small 17–base pair section of DNA from each species. For example, one probe might match cow DNA, and another might match goose DNA. By looking at which probes detect their complementary DNA, researchers can tell which species are present in the sample.
Employing the same manufacturing technology that is used to build microprocessors, thousands or millions of probes can be synthesized simultaneously. In fact, the surface of a microarray looks like a giant checkerboard, with each square holding one unique type of probe. Each probe is built one layer at a time, one base stacked on top of another, like checkers, until a full-length probe is created (Fig. 3). The finished microarray is about the size of a thumbnail, with approximately 97,000 squares, called “features.” Each feature, which contains a genetic marker for one of the 33 different animal species, is about 26 microns wide (a human hair is about 50 microns wide).
With a probe array designed to detect an animal’s DNA, the next step is to homogenize and extract that DNA from a food sample. With primers provided in the FoodExpert-ID kit, a single polymerase chain reaction (PCR) is used to amplify cytb gene sequences from all detectable species. Using additional buffers included in the kit, the amplified cytb DNA is then transcribed into RNA, and is simultaneously “marked” with a fluorescent rUTP-fluorescein ribonucleotide. Essentially, the RNA is tagged with a fluorescent label, similar to how animals in the wild are tagged with tracking devices.
The labeled RNA is then injected into the microarray, where it is “washed over,” or hybridized to the probes on the array surface. If the RNA from the food sample matches the DNA on the microarray, the strands of RNA will pair with their partner probes (Fig. 4). To detect a match, scientists place the array into a scanning device, where a laser is used to illuminate the fluorescently tagged RNA, causing the squares containing a matched DNA probe to light up (Fig. 5). After the array is scanned, the image is transfered into a computer, where a data file is generated and then analyzed by software to determine if a species is present in or absent from the original sample. Fig. 6 shows how the labeled RNA is injected into the microarray, and Fig. 7 shows a scanned image.
In actual practice, there are more than 80,000 different probes used to detect 33 species of animals on every microarray. The resulting picture is complex, but when analyzed, researchers can see the different levels of fluorescence coming from the individual squares or probe locations (Fig. 8). By analyzing the fluorescence intensity, they can determine what animal species are present in a sample. If the probes for a specific animal DNA light up, the results are clear and simple—the animal is present in the sample. If they do not light up, the results are equally clear—what you thought you were eating isn’t what’s on your plate. From start to finish—extracting DNA to determining food composition—the process routinely takes about 48 hr.
--- PAGE BREAK ---

Performance Criteria
Like other diagnostic devices, the microarray’s performance is bound by measures of specificity and sensitivity. Inclusive and exclusive specificity of the assay has been validated for each of the 33 species detected on the array. At least three additional species are detectable (Virginian deer, American eel, and Pacific bonito), but their specificity has not yet been fully validated. The sensitivity of the assay is proven at 5%, meaning that animal species can routinely be identified from a vegetable matrix when constituting at least 5% of the total sample weight.

In tests of actual food products of certified origin, the microarray has determined the species present with 90% accuracy—even for foods which have been cooked, smoked, salted, deep-frozen, or canned. However, common food additives of animal origin, which do not explicitly contain DNA, such as milk proteins and egg white, are not detectable by the microarray, since the system requires DNA to be present to ascertain species composition.
To guarantee correct interpretation of the microarray probe data, the Food-Expert-ID kit contains four controls which are processed along with the sample. A hybridization control is used to validate hybridization and the DNA microarray reading steps for each test. Internal controls detect protocol problems resulting from sample preparation or human error, such as the presence of PCR inhibitors or omission of a reagent. A negative control consists of blank reagents and checks for environmental contamination. And a positive control, consisting of a reference sample (e.g., beef DNA) ensures that the complete test environment is functioning properly.
By including each of these four controls, the interpreted results can be relied on to draw highly accurate food composition conclusions.
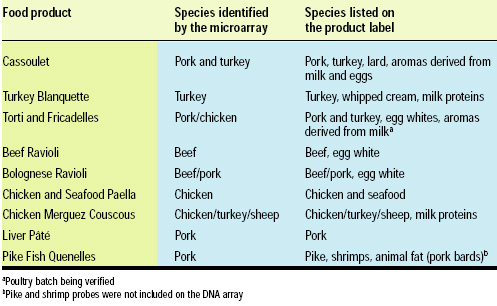
bioMérieux collaborated with Atlangène Applications, a leading French DNA analysis laboratory, to test a number of off-the-shelf canned foods for agreement with the species identified by the microarray. The findings, shown in Table 2, showed very good correlation with the labeling, validating the utility and reliability of the microarray for determining food composition. While finding pork to be present in pike quenelles was at first surprising, its presence was confirmed by the manufacturer.
The Way Ahead
The FoodExpert-ID uses the same Affymetrix technology that has revolutionized genetic studies in laboratories around the world, demonstrating the potential for use of microarrays in food testing and quality control. As microarray technology continues to increase in information density, becoming more cost efficient, we anticipate that the technology will find many other uses where it can be advantageous to look at life at the molecular level.
The fabrication parallels between microarray technology and semiconductor technology afford microarrays the same degree of potential that allowed computer microprocessors to progress from an obscure feat of engineering to the standard inner workings of a child’s toy. In the food industry alone, microarrays have the potential to distinguish fish species at the Tokyo fish market, validate Cabernet Sauvignon wine, or even figure out what exactly is in mom’s meatloaf.
The author is Director of Business Development, Molecular Diagnostics, Affymetrix, Inc., 3380 Central Expressway, Santa Clara, CA 95051, [email protected].
