Nanosensors Identify Pathogens in Food
Biosensors similar to pregnancy test kits are being developed for the rapid, reliable, yet inexpensive identification and quantification of pathogenic organisms. Nanotechnology plays a major role.
In general, current technology can provide all of the data necessary to ensure the safety of our food, if neither time nor cost were a concern. For example, standard microbiological assays can provide the ultimate sensitivity and identify the presence of a single cell in a given food sample. However, these assays suffer from long incubation steps and therefore provide results only after 24–48 hr or even longer. They also suffer from relatively complex procedures that require handling by trained technicians in a laboratory setting.
More recently, lateral flow assays—the technology used in pregnancy test kits, for example—have been used to substitute for some of the complex sample handling. They are used to detect pathogens after a 24- to 48-hr incubation, clearly ameliorating the complexity of the overall procedure. These tests are based on antibodies that can determine the presence of pathogens such as Campylobacter jejuni, Salmonella, Listeria monocytogenes, and Escherichia coli O157 with a 10- to 20-min assay. The more-cutting-edge molecular biological assays employing polymerase chain reaction (PCR) that identify pathogens via their nucleic acid sequences can also reach detection limits of a few cells per food sample sometimes within 6 hr. However, they are still very complex and expensive techniques that need to be performed in a highly specialized laboratory.
Two Technologies Pursued
Our laboratory is focusing on the development of rapid, inexpensive, easy-to-use, portable, yet highly sensitive and reliable biosensors that can identify and quantify pathogenic organisms. The ideal device will be a “black box” the size of a large cell phone with a sample-inlet hole, an on/off button, and a digital readout.
To reach this ideal device, we are developing two different technologies that use pathogens such as Bacillus anthracis (Hartley and Baeumner, 2003; Baeumner et al., 2004), Cryptosporidium parvum (Esch et al., 2001), Dengue virus (Baeumner et al., 2002; Zaytseva et al., 2004), and general E. coli (Baeumner et al., 2003) as model analytes. The two technologies are a lateral flow RNA assay and a bioanalytical microsystem.
The bioanalytical microsystem, fabricated using nanotechnology, contains a microfluidic biosensor and will ultimately bear all of the desired characteristics of the black box type of pathogen sensor.
We consider the lateral flow RNA assay a “precursor biosensor,” since we are investigating and establishing in this format the molecular biological strategies that will subsequently be used in the more sophisticated bioanalytical microsystem. Its simplicity and low cost allow a very rapid analysis of a variety of different conditions, and it is therefore ideal for the optimization process. In fact, we have generated a universal nucleic acid lateral flow assay that is ideal for prototyping and will be described further below.
We have been collaborating extensively with Richard Durst of Cornell University, who was the first to show the exceptional usefulness of liposomes as analytical reagents and who is focusing today on antibody- and receptor-based assays; and we are working with Richard Montagna of Innovative Biotechnologies International, Inc., to expand our technology and prove its adaptability to a variety of different analytes.
--- PAGE BREAK ---
• Lateral Flow RNA Assay. The simple lateral flow assay achieves detection limits of 1.5–10 fmol (1.5–10 nM). The time required per assay is about 10–15 min, as soon as RNA from the pathogen is available. No special equipment, training, or laboratory setting is required to perform the assay. If low numbers of pathogens (as low as 5 organisms/mL) have to be detected, sample preparation adds about 3 hr to the procedure, which includes RNA purification and amplification using the isothermal nucleic acid sequence–based amplification (NASBA) reaction.
This assay has been able to highly specifically detect 5 viable C. parvum oocysts, 40 viable E. coli cells, and 10 B. anthracis spores. While this is not a portable assay because of the included sample preparation steps, device requirements (an added water bath and small centrifuge) are minimal, and total assay times of only 4 hr are quite acceptable.
• Bioanalytical Microsystem. Our more sophisticated bioanalytical microsystem has several advantages over the simple lateral flow assay: (1) the detection limit is lowered by at least one order of magnitude to 0.1 fmol per assay, which also directly translates into a faster overall assay since a shorter NASBA reaction is required; (2) the sample preparation steps are integrated into the device so that no sample handling is required by the user; and (3) the overall assay time from raw sample to result can be reduced to less than 1 hr.
• Benefits of Both. How can our lateral flow assay and the bioanalytical microsystem be so sensitive, specific, yet simple and fast? There are four reasons:
1. We take advantage of the high specificity of the genome for our detection approach. In addition, instead of using DNA as our analyte, we detect messenger RNA (mRNA), which is only present when the organism is alive. Thus, a dead pathogen will not produce false positive signals.
2. We use liposomes as our signal-generation and signal-amplification system. Liposomes are small spheres made of a phospholipid bilayer entrapping an aqueous cavity. Liposomes are typically quite small—they are in fact nanoparticles, made with a diameter of 200 nm. This large (on the nanoscale) vesicle can entrap millions of molecules of dye or electrochemical marker and therefore provides a million-fold signal amplification.
3. We use standard photolithography and softlithography processes to fabricate the bioanalytical microsystem, with the goal of using only designs that are simple. While this makes the development itself more complicated, it guarantees that the ultimate device will be a simple-to-use biosensor.
4. The bioanalytical microsystem integrates detection and sample preparation on one chip, so no user handling is required and the entire procedure can be carried out in one portable device. The design for this is modular, in that the sample preparation steps—cell lysis/disruption (Dhawan and Baeumner, 2002), RNA purification, and RNA amplification—are carried out in separate modules. The fourth module is the microfluidic biosensor (Kwakye and Baeumner, 2003).
--- PAGE BREAK ---
How the Biosensor Technology Works
As mentioned above, in our biosensors pathogenic organisms are identified via their intracellular mRNA. To do this, it is, unfortunately, not sufficient to simply dip the biosensor into a sample containing the pathogen. Instead, a number of analytical operations are required (Fig. 1). Thus, in addition to the detection of the RNA itself, cell disruption, RNA purification, and amplification via NASBA are needed. In the case of the lateral flow assay, these steps are carried out in the lab using a water bath and small centrifuge. In the case of the bioanalytical microsystem, each step is realized in a separate module that taken together, form the microsystem.
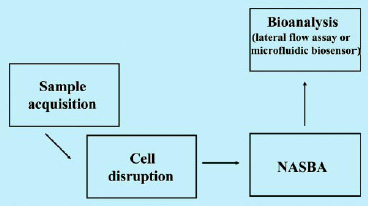
Figs. 2 and 3 show the principle of the lateral flow assay. The RNA from the pathogen is detected via an RNA/DNA hybridization event. Thus, we have designed two different DNA probes that can bind to two different portions of the target RNA, forming a sandwich (the two DNA probes are the slices of bread and the target RNA is the cheese). One of the two DNA probes (the reporter probe) is immobilized on the outside of the liposomes, and the other (the capture probe) is immobilized on a distinct location of a nitrocellulose membrane (in the detection zone).
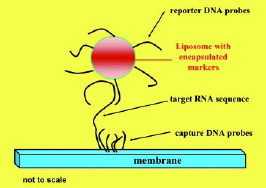
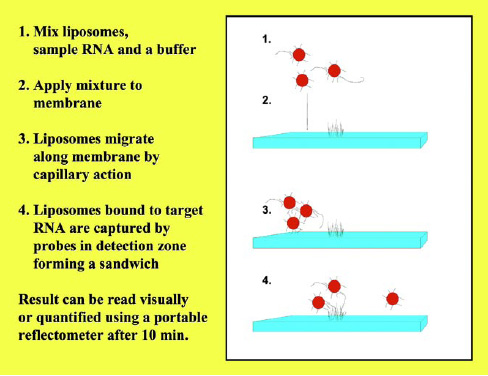
In the assay, the liposomes and the sample are mixed and applied to the membrane, and the mixture migrates along the membrane strip by capillary action. When the mixture reaches the detection zone, the capture probes bind to the target RNA. Thus, in the detection zone, the presence of the target RNA molecules can directly be quantified by the amount of liposomes present. Since the liposomes entrap red dye molecules in this assay, the signal can be observed either visually or, more quantitatively, via a portable reflectometer that measures the reflectance of the liposomes in the detection zone. As mentioned earlier, detection limits in the low nM range can be obtained with this simple 10-min assay.
In the case of the bioanalytical microsystem, not only the detection module but also the sample preparation modules have to be developed. All of the micromodules are designed using computer aided design software, nanofabricated in a silicon wafer by photolithography, and finally molded into a polydimethyl siloxane (PDMS) elastomer by softlithography. The same procedure is applied to all of the different modules to ensure their compatibility for later assembly, i.e. all modules are made in PDMS with similar feature sizes.
To connect the modules to the outside (macro) world, a Plexiglas housing provides fluid and electrical contacts. Fig. 4 is an example of the assembled microfluidic biosensor module. In addition to the modules, active and passive elements are integrated to develop a fully functioning and independent bioanalytical microsystem that can be operated by a small battery. These elements include microheaters, micromixers, and micropumps.

Fig. 5 is an example of a possible micromixer. A “honeycomb structure” created in the PDMS elastomer promotes the passive mixing of two solutions by causing them to flow through the intricate design.
--- PAGE BREAK ---

The same detection principle is used in the detection module of the bioanalytical microsystem as in the lateral flow assay. The only difference is that in the bioanalytical microsystem the liposomes entrap electrochemically active markers instead of dye molecules. Thus, the signal generated is not visually detectable, but can be reported via oxidation and reduction reactions on an electrode we developed. Experiments are underway that integrate ultramicroelectrode arrays (IDUAs) into the microfluidic biosensor module (Min and Baeumner, 2004). This approach is extremely interesting because of the lower detection limit offered by electrochemical detection and the simplicity of the device needed to operate the IDUAs. Fig. 6 shows the IDUAs integrated with the PDMS microchannel. The final “cell phone-sized” bioanalytical microsystem will contain the IDUAs as a transducer and will be operated by a micropotentiostat.
What’s Next
We have also developed a universal biosensor that can be made specific for any pathogenic organism of interest within a few minutes with no special equipment and skills. It detects pathogens on the basis of their nucleic acid sequences. The biosensor is based on our specific-pathogen lateral flow biosensors described above (Baeumner et al., 2004b). We generated a universal membrane and a universal liposome that are made specific within a simple 10-min incubation step. These biosensors reach the same detection limits and similar dynamic ranges as their specific counterparts do. Thus, developing biosensors for new pathogenic organisms has become a very simple and routine task in our lab. The next step is the integration of this universal approach into the bioanalytical microsystem.
The lateral flow assay is a technology ready for commercialization today. The bioanalytical microsystem will not be available for commercialization for 2–4 more years of development. Nanotechnology is critical in the development of new bioanalytical tools that will help achieving the goal of building the ultimate pathogen sensor for the consumer and first responder. Our approach, in which modules for sample preparation and detection are integrated on the same platform, allows the design of portable and easy-to-use analytical tools. Our liposomes are nanovesicles that serve as the ideal bioanalytical signal-generation reagent, since instantaneous and million-fold amplification can be obtained.
Nanotechnology will also be important for the development of novel materials that will make sample preparation from complex food matrices less challenging. Research is needed to develop nanomaterials that help integrate the nano- and micro-world with the required macro-world of food analysis. This is especially important considering that typical nano- and microsensors can analyze only tiny volumes, often less than 1 µL, which cannot provide information representative of an entire food sample that might be several hundred grams or several hundred milliliters in size. Thus, “smart” novel materials can be developed that can extract pathogens directly on the biochip or that can be added to the sample prior to bioanalysis to enable the analysis of sample sizes greater than 1 g and 1 mL.
Thus, it is critical to apply nanotechnology to the development of new materials and new techniques that can be utilized for the development of suitable sample preparation steps, such as extraction, concentration, and isolation. Combining this with rapid, portable, highly sensitive, specific, yet inexpensive detection technology will allow us to overcome challenges we encounter today in pathogen detection.
The author is Assistant Professor of Biotechnology, Bioanalytical Microsystems and Biosensor Laboratory, Dept. of Biological & Environmental Engineering, 318 Riley-Robb Hall, Cornell University, Ithaca, NY 14853, [email protected].
References
Baeumner, A.J., Schlesinger, N.A., Slutzki, N.S., Romano, J., Lee, E.M., and Montagna, R.A. 2002. A biosensor for Dengue virus detection: Sensitive, rapid and serotype specific” Anal. Chem. 74: 1442-1448.
Baeumner, A.J., Cohen, R.N., Miksic, V., and Min, J.H. 2003. RNA biosensor for the rapid detection of viable Escherichia coli in drinking water. Biosensors Bioelectronics 8: 405-419.
Baeumner, A.J., Leonard, B., McElwee, J., and Montagna, R.A. 2004a. A rapid biosensor for viable B. anthracis spores. Anal. Bioanal. Chem. In press.
Baeumner, A.J., Pretz, J., and Fang, S. 2004b. A universal nucleic acid sequence biosensor with nanomolar detection limits. Anal. Chem. 76: 888-894.
Dhawan, M.D., Wise, F., and Baeumner, A.J. 2002. Development of a laser-induced cell lysis system. Anal. Bioanal. Chem. 374: 421-426.
Esch, M.B., Baeumner, A.J., and Durst, R.A. 2001. Detection of Cryptosporidium parvum using oligonucleotide-tagged liposomes in a competitive assay format. Anal. Chem. 73: 3162-3167.
Hartley, H.A. and Baeumner, A.J. 2003. Biosensor for the specific detection of a single viable B. anthracis spore. Anal. Bioanal. Chem. 376: 319-327.
Kwakye, S. and Baeumner, A. 2003. A microfluidic biosensor based on nucleic acid sequence recognition. Anal. Bioanal. Chem. 376: 1062-1068.
Min, J.-H. and Baeumner, A.J. 2004. Characterization and optimization of interdigitated ultramicroelectrode arrays as electrochemical biosensor transducers. Electroanalysis 16: 724-729.
Zaytseva, N., Montagna, R.A., Lee, E.M., and Baeumner, A.J. 2004. Multi-analyte single-membrane biosensor for serotype specific detection of Dengue virus. Anal. Bioanal. Chem. In press.
