Beyond Nutrition The Impact of Food on Genes
Nutrigenomics—understanding how nutrients affect genes—will enable foods to be developed that can be used to prevent and treat disease.
The food we eat is composed of complex mixtures of chemicals, many of which are biologically active. Whether nutritive (vitamins, minerals, fatty acids) or non-nutritive (small-molecule “nutraceuticals” such as phytochemicals or other metabolites), these bioactive substances act as dietary signals that manipulate our highly organized, tightly regulated DNA. While consumers mainly focus on taste, convenience, and price, arguably the most important effects of food occur at the molecular level, generating significant effects on disease states in humans, whether good or bad. The science that studies the effect of dietary bioactive compounds on gene expression is called nutrigenomics.

The Potential of Nutrigenomics
Regulation of gene expression is actually a primitive adaptive mechanism for survival. Organisms can sense the absence or presence of metabolites and respond to nutrient deficiency or excess by increasing or decreasing production of cellular proteins necessary for a myriad of functions. During fermentation, including brewing, yeast strains will alter their metabolic functions and can actually be manipulated through environmental conditions (concentration of sugars, addition of sterols or unsaturated fatty acids, control of oxygenation) to enable a stress response which alters gene expression (Brosnan et al., 2000).
With the substantial data that support the dramatic effect of nutrients on gene expression in cultured microorganisms, it is not surprising that most foods we consume contain bioactives that are capable of modulating gene expression in humans. This is very important because as the science of nutrigenomics continues to evolve, it will reveal not only which foods can have a direct impact on our genes, but also the mechanisms as to how and why. The ultimate goal is to be able to prevent, and potentially even treat, disease through targeted nutrition.
In 1996, Ghai and coworkers filed a seminal patent which highlighted the potential for development of foods or supplements that could alter the expression of genes associated with human diseases (Ghai et al., 1999). They demonstrated that certain flavonoids found in citrus peel enhanced expression of a gene involved in the human body’s natural defense against cancer. This was the beginning of the study of the relationship of food, as nutrition, and our genes, now called nutrigenomics.
Nutrigenomics is slowly making its way into the mainstream food industry. The IFT Expert Panel Report on Functional Foods (Clydesdale, 2004; IFT, 2005) described nutrigenomics, proteomics, and metabolomics as “the three new disciplines that will contribute to the rapid development of functional foods [to] identify the biological basis by which food components promote health and wellness.” As defined in the report, “nutrigenomics” represents the interaction of dietary components and genes, whereas “proteomics” is the study of the proteins encoded by the genes and “metabolomics” is the systems biology approach for measuring the outcomes of the potential changes from genomics and proteomics.
The current research and potential for nutrigenomics have been highlighted in numerous recent articles and reviews, including Gillies (2003), Levi and Sanderson (2004), Müller and Kersten (2003), Kauwell (2005), and Fogg-Johnson and Kaput (2003).
Genetic Effects of Food
It has been recognized for many years that diet is an important factor in the onset or proliferation of human disease, as well as in prevention. Chemicals found in food interact with biochemical pathways at the molecular level—for example, to elicit allergic reactions or alter (potentially increase or decrease) the levels of biomarkers, such as blood/sugar, cholesterol, and various proteins.
Dannenberg and Reidenberg (1994) suggested that dietary fats act as drugs. They detailed the pharmacological effects associated with numerous fatty acids that are part of our daily diet, including modification of blood pressure, cholesterol levels, risk of carcinogenesis, and levels of pre-inflammatory chemicals. Since we don’t consume only individual nutrients but instead eat the rather complicated composites otherwise known as food, the situation is much more complex. While most measurements of the impact of food on human biochemistry utilize quantification of biomarkers, evidence is mounting that, in many cases, modulation of gene expression by bioactives is an important mechanism of action. If the genes are controlling the output (i.e., biomarkers), it makes sense to measure the driver of the reactions.
--- PAGE BREAK ---
While researchers are exploring methods to identify bioactives that alter gene expression in humans, the food we eat is already altering expression of our genes. Folic acid was identified in early research as an important regulator of gene expression. Folate provides the precursors for purine and thymidylate syntheses and for methylation of DNA, and therefore plays a direct role in regulation of gene expression. Undermethylation of DNA—a prerequisite for proliferation of cancer cells—has been detected in humans fed a folate-deficient diet.
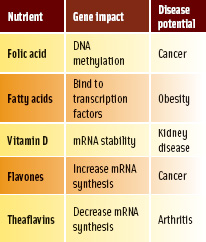 Nearly all evidence for the impact of nutrients on gene expression is derived from research using animal or human cells in culture. Nutrient/gene/disease interactions have been documented for several common food substances. Vitamins (tocopherol, biotin), minerals (zinc), and phytochemicals (flavones, catechin) have all been demonstrated to alter gene expression in human cells in culture. Cells in culture offer the opportunity to elucidate the mechanism by which food substances can alter gene expression. Several methods have been identified whereby nutrients interfere with gene expression. These include direct involvement in DNA synthesis (biotin metabolites), DNA methylation (folic acid), and alteration of mRNA stability (vitamin D) (Table 1).
Nearly all evidence for the impact of nutrients on gene expression is derived from research using animal or human cells in culture. Nutrient/gene/disease interactions have been documented for several common food substances. Vitamins (tocopherol, biotin), minerals (zinc), and phytochemicals (flavones, catechin) have all been demonstrated to alter gene expression in human cells in culture. Cells in culture offer the opportunity to elucidate the mechanism by which food substances can alter gene expression. Several methods have been identified whereby nutrients interfere with gene expression. These include direct involvement in DNA synthesis (biotin metabolites), DNA methylation (folic acid), and alteration of mRNA stability (vitamin D) (Table 1).
How Nutrients Affect Genes
A key mechanism of action involving nutrient–gene interaction is the ability and necessity of some nutrients to act as ligands and bind with transcription factors. Transcription factors bind to specific DNA sequences in the promoter region of specific genes, thereby either enhancing or suppressing gene expression. Binding of a nutrient to a transcription factor will affect the ability of a transcription factor to bind to DNA. For example, peroxisome proliferation–activated receptors (PPARs), a family of steroidal, nuclear transcription factors, affect expression of genes associated with fatty acid metabolism, energy balance, eico-sanoid signaling, and tumorigenesis (Kliewer et al., 2001). In an intricate, multi-step process, fatty acids (e.g., omega-3s and omega-6s from salmon) bind to the PPAR, which then in turn binds to another transcription factor called retinoid X receptor (RXR) that is activated only when vitamin A derivatives (such as those from carrots) bind (Ouamrane et al., 2003). Gene expression results from the fatty acid/PPAR–retinoid/RXR complex that directly binds to the DNA.
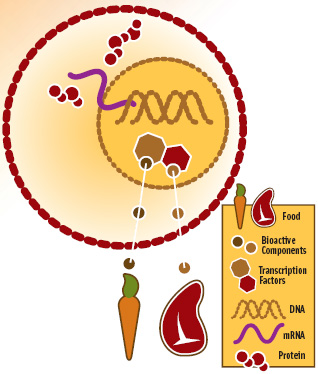 Figure 1 is a simplistic pictorial illustrating that some of the food we consume is made up of bioactive ingredients that pass through our cells into the nucleus, where they act as ligands to directly bind to DNA to effect transcription of RNA, and ultimately translation of proteins. In the case of retinoids from carrots and fatty acids from salmon, they bind to specific genes via a PPAR complex, resulting in, for example, reduction of fatty acid synthesis and increase in fatty acid oxidation.
Figure 1 is a simplistic pictorial illustrating that some of the food we consume is made up of bioactive ingredients that pass through our cells into the nucleus, where they act as ligands to directly bind to DNA to effect transcription of RNA, and ultimately translation of proteins. In the case of retinoids from carrots and fatty acids from salmon, they bind to specific genes via a PPAR complex, resulting in, for example, reduction of fatty acid synthesis and increase in fatty acid oxidation.
The number of nuclear receptors capable of binding fatty acids seems to increase daily. The list of genes regulated by fatty acids is extensive (Pegorier et al., 2004). It includes genes involved in fatty acid transport, oxidation of fatty acids, desaturation of fatty acids, glycolysis, lipogenesis, and lipoprotein metabolism. Studies in mice have demonstrated that fatty acids can have an impact on expression of a large number of genes. Berger et al. (2002) fed mice diets rich in omega-6 or omega-3 fatty acids that exceeded nutritional requirements. When expression of 12,000 genes was analyzed using DNA microarrays, more than 300 genes differed in expression levels.
Moving from Lab Studies to Humans
Every cell has DNA. It is difficult to imagine how dietary substances could affect the DNA in all the cells in the human body. While the food we eat comes in direct contact with cells of the digestive tract, there is no direct contact of the food we eat with other important organs. For a food substance to affect a gene in the liver, it must be transported into and through the bloodstream (presumably without being metabolized), reach the liver, enter the liver cytoplasm, and then enter the liver cell nucleus.
--- PAGE BREAK ---
Results of animal studies are encouraging. It has been demonstrated in rats that modification of zinc deficiency by zinc supplementation modifies expression of many genes. Fong et al. (2005) demonstrated that zinc-deficient rats administered intra-gastric zinc had an 80% reduction in mRNA of cyclo-oxygenase-2 (COX-2), a key enzyme involved in inflammation, after only 8 hr. Supplementation with vitamin E can also modulate gene expression in rat livers.
In addition, caloric restriction has been shown to result in increased gene expression of insulin-related genes in rats (Zhu et al., 2005). It has also been observed that gene expression is altered in the livers of pigs fed casein or soy protein. Zeng et al. (2003) observed increased levels of gene transcription for several apoptosis- related genes in the livers of mice fed selenium-enriched broccoli. High-fat diets also result in increased expression of genes important for fat oxidation.
Matsui et al. (2005) demonstrated the effect of cocoa on gene expression in rats. Rats fed a diet containing 12.5% cocoa powder for 21 days had lower body weight and less white adipose tissue than controls. They had lower expression of genes for fatty acid synthesis in the liver and white adipose tissue and decreased expression of fatty acid transport genes in white adipose tissue.
It is anticipated that alterations in gene expression should be associated with disease states. Zeng et al. (2003) and Fong et al. (2005) examined various molecular and cellular markers related to cancer. Treatment with selenium and zinc, for example, suggested potent anti-cancer effects.
To understand the full impact of nutritional factors on gene expression in humans it will ultimately be necessary to complete clinical studies. While human cell data are compelling, just because a bioactive is shown to have an effect on gene expression in cells in culture—or even in animal models, for that matter—doesn’t necessarily translate to having similar effects in humans.
Unfortunately, studies with humans are limited. Cao and Cousins (2000) examined gene expression of a gene encoding a zinc-binding protein (metallothionein), using blood samples from 16 men. Supplemented subjects were given the recommended dietary allowance for zinc for 10 days. After two days, significantly higher mRNA levels of the gene for zinc-binding protein could be detected in blood cells, with elevated mRNA levels maintained throughout the 10 days of zinc supplementation. Cameron-Smith et al. (2003) demonstrated that a short-term, high-fat diet increases both lipid metabolism and gene expression in human muscle tissue. Increased gene expression was observed for genes involved in fatty acid transport and β-oxidation in individuals fed a high-fat rather than a high-carbohydrate diet.
Targeting Inflammation
Nutrigenomics may seem more like a basic science rooted in genetics, but in the context of food it is really an applied science. Food science is merely trying to apply the many years’ worth of research in genomics (such as proteomics, metabolomics, pharmacogenomics, molecular biology, etc.) that is discovering and elucidating the pathways and mechanisms of action of the more than 20,000 genes that have been identified based on recent sequencing of the human genome.
Nutrigenomics seeks to understand if and how foods and plant extracts can affect the expression of genes in well-known and characterized pathways responsible for the complex, chronic diseases, such as cardiovascular disease (primarily heart disease and stroke), cancer, diabetes, and arthritis.
An important area for identification of bioactives is inflammation, which leads to accelerated development of a multitude of chronic diseases. The direct or indirect involvement of chronic inflammation in several important human diseases (such as cardiovascular disease, cancers, diabetes, arthritis, immune dysfunction, and joint pain) and the dietary causes of inflammation (saturated fats and trans fats) were reviewed by Finley (2004).
--- PAGE BREAK ---
Naturally occurring nutraceuticals, specifically antioxidant bioactives, such as plant phenols, vitamins, carotenoids, and terpenoids, have been shown to have significant beneficial effects for health promotion by reducing the process of sustained inflammation that accompanies chronic disease. Several genes directly involved in inflammation include COX-2, tumor necrosis factor α (TNF-α), interleukin-1 (IL-1), phospholipase A2, 5-lipoxygenase (LOX), and inducible nitric oxide synthase (iNOS). Figure 2 shows the highly complicated pro-inflammatory metabolic pathways that result from the body’s breakdown of omega-6 fatty acids. Down-regulation of the genes which code for the COX and LOX enzymes to decrease production of prostaglandins or leukotrienes may contribute to prevention or treatment of the associated health conditions. Omega-3s, on the other hand, such as EPA and DHA, are competitive inhibitors of both COX and LOX, thereby having anti-inflammatory effects.
Subbaramaiah et al. (2001) screened nearly 1,000 plant extracts in an effort to identify bioactives capable of modulating expression of the COX- 2 gene. Using human cell cultures, researchers have identified several food substances that are able to down-regulate expression of one or more of these genes, including curcumin from turmeric, resveratrol from grapes, catechins from green tea, theaflavins from black tea, and vitamin E.
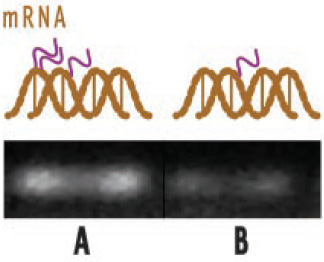 For example, Figure 3 illustrates the effect of theaflavin from black tea on expression of COX-2, a gene involved in inflammation and pain. It shows reverse transcription–polymerase chain reaction products showing expression of RNA by electrophoresis. The brighter the white line, the greater the expression of the gene involved. The left side of the figure shows that with no theaflavin, COX-2 is activated in tumor cells, producing substantial mRNA. The right side shows that treatment of cells with theaflavins results in reduction in gene expression and therefore lower mRNA. Ultimately, this results in lower production of COX-2, a key enzyme linked to prostaglandin synthesis (see Figure. 2).
For example, Figure 3 illustrates the effect of theaflavin from black tea on expression of COX-2, a gene involved in inflammation and pain. It shows reverse transcription–polymerase chain reaction products showing expression of RNA by electrophoresis. The brighter the white line, the greater the expression of the gene involved. The left side of the figure shows that with no theaflavin, COX-2 is activated in tumor cells, producing substantial mRNA. The right side shows that treatment of cells with theaflavins results in reduction in gene expression and therefore lower mRNA. Ultimately, this results in lower production of COX-2, a key enzyme linked to prostaglandin synthesis (see Figure. 2).
Human Validation is Key
Human clinical studies are needed to make a cause-and-effect relationship between the food affecting gene expression and the disease state. Carefully designed double-blind, placebo-controlled clinical studies—with biomarkers clearly defined and measured and personal qualitative life-quality issues recorded—are needed for credibility. Epidemiological studies analyzed to describe relationships among populations would also be extremely useful to support the science. These types of studies lay the foundation for exploring the multitude of reasons and factors why measurable benefits are achieved with the foods (or supplements) being studied.
Thanks to traditional Chinese medicine and Indian Ayurvedic medicine, we have generations of data that show a beneficial cause-and-effect relationship between many plants (including herbs) and foods against human disease. These cultural histories are rich with associations between health-promoting foods/plants and specific diseases. Again, nutrigenomics becomes the means to confirm the mechanism of action to describe why something “works.”
Having a history of consumption is also obviously a benefit in building a case for safety, in addition to the basic premise that plants and foods, as the starting material, have potentially favorable regulatory status, e.g., as Generally Recognized As Safe (GRAS) substances. Also, an ingredient can be created, and the GRAS self-affirmation process streamlined, as long as toxicological testing deems that the product is not life-threatening for the intended use and application at the proposed dose.
While studies often evaluate the effect of individual genes, it is important to realize the complexity of chronic diseases which involve multiple genes and metabolic and signaling pathways. To truly understand the biology of processes directed by genes, researchers need to simultaneously study the genetic influences as well as some ultimate positive quantitative biological response, such as a biomarker. For any functional ingredient, whether discovered through genomics or not, a positive health effect must be elicited and potentially sustained with some form of measurable outcome.
Future of Nutrigenomics
As science has begun to identify genes associated with human disease, it is not surprising that chemicals in food have been demonstrated to alter expression of these genes. It is not difficult to imagine that nutrigenomics will play an important role in addressing expression of genes responsible for single-gene diseases. There are many examples where a single-gene mutation is responsible for a disease, but in most cases these are diseases that affect a very small proportion of the human population.
--- PAGE BREAK ---
The real challenge for nutrigenomics is to target the genes involved in the major human disease states, such as cancer, heart disease, obesity, and arthritis, where these health problems merely represent generic names for a multitude of actual diseases, with multiple genes that affect disease susceptibility.
Many of the genes involved in these polygenic, chronic diseases have not yet been identified. For example, it is possible that a substance may turn on some genes involved in inflammation, while simultaneously turning offother genes involved in inflammation.
Since many diseases of interest are chronic, and since the primary focus of nutraceuticals is prevention, rather than treatment, it is necessary to have endpoints that are meaningful to consumers. Development of successful nutrigenomics businesses will require continued development of meaningful biomarkers and completion of innovative clinical trials that monitor biomarkers. For products targeting prevention, it will be necessary to have convenient, inexpensive ways for consumers to verify the effectiveness of nutrigenomics products.
Nutrigenetics Focuses on Genetic Make-Up
Nutrigenomics (Gilles, 2003; German, 2005), in essence, is the interaction of nutritional components (especially bioactives) on human genes. It effects the change in expression of genes as a consequence of ingestion of a bioactive. Nutrigenetics, a subset of nutrigenomics, attempts to identify human allelic variations that respond differently to bioactives. Nutrigenetics recognizes an individual’s hereditary predisposition to disease based on his or her genetic make-up. Ultimately, via a nutrigenetic approach, recommendations of dietary supplements and whole foods will be based on an individual’s genetic background.
There are already fascinating, and hopefully prophetic, examples of nutrigenetics, in which genetic variation between individuals influences dietary responses. Ordovas et al. (2002) demonstrated how a single-point mutation in the APOA1 gene alters how an individual responds to the effect of polyunsaturated fatty acids on HDL cholesterol levels. It has also been demonstrated that polymorphisms are important in the ability of fish oil to reduce tumor necrosis factor-α, a key mediator of inflammation. Nonetheless, there are significant technical hurdles that must be overcome before we will see the benefits of nutrigenetics.
Nutrigenetics is dependent on diagnostics. Fogg-Johnson and Kaput (2003) identified several of the technical issues with diagnostics that suggest that we are many years from development of meaningful low-cost diagnostic tests that can be used to predict personalized nutritional requirements. Marshall (2004)’s report of inconsistent results from comparison among commercial DNA microarrays underscores the need to validate any diagnostic test against biomarkers in a human clinical setting before commercial diagnostic tests are mass produced.
Author Hirsch is Director, Product Development, and Author Evans is CEO, WellGen, Inc., 63 Dudley Rd., New Brunswick, NJ 08901. Send reprint requests to author Hirsch ( [email protected] ).
www.ift.org
Members Only: Read more about nutrigenomics online at www.ift.org. Type in the keyword in our search box in the upper left side of our home page.
References
Berger, A., Mutch, D.M., German, B., and Roberts, M.A. 2002. Dietary eff ects of arachidonate-rich fungal oil and fi sh oil on murine hepatic and hippocampal gene expression. Lipids Health Dis. 1: 1-23.
Brosnan, M.P. Donnelly, D. James, T.C., and Bond, U. 2000. The stress response is repressed during fermentation in brewery strains of yeast. J. Appl. Microbiol. 88: 746-755.
Cameron-Smith, D. Burke, L.M., Angus, D., Tunstall, R., Cox, G., Bonen, A., Hawley, J., and Hargreaves, M. 2003. A short term, high fat diet upregulates lipid metabolism and gene expression in human skeletal muscle. Am. J. Clin. Nutr. 77: 313-318.
Cao, J. and Cousins, R.J. 2000. Metallothionein mRNA in monocytes and peripheral blood mononuclear cells and in cells from dried blood spots increases after zinc supplementation of men. J. Nutr. 130: 2180-2187.
Clydesdale, F. 2004. Functional foods: Opportunities & challenges. Food Technol. 58(12): 35-40.
Dannenberg, A. and Reidenberg, M.D. 1994. Dietary fatty acids are also drugs. Clin. Pharmacol. Ther. 55: 5-9
Finley, J. 2004. Phenolic antioxidants and prevention of chronic infl ammation. Food Technol. 58(11): 42-46.
Fogg-Johnson, N. and Kaput, J. 2003. Nutrigenomics: an emerging scientifi c discipline. Food Technol. 57(4): 60-67.
Fong, L.Y.Y., Zhang, L., Jiang, Y., and Farber, J.L. 2005. Dietary zinc modulation of COX-2 expression and lingual and esophageal carcinogenesis in rats. J. Natl. Cancer Inst. 97: 40-50.
German, J.B. 2005. Genetic dietetics: Nutrigenomics and the future of dietetics practice. J. Am. Dietet. Assn. 105: 530-531.
Ghai, G., Boyd, C., Csiszar, K., Ho, C.T., and Rosen, R.T. 1999. Methods of screening foods for nutraceuticals. U.S. patent 5,955,269.
Gillies, P.J. 2003. Nutrigenomics: the rubicon of molecular nutrition. J. Am. Dietet. Assn. 103(12, Suppl. 2): S50-S55.
IFT. 2005. IFT Expert Panel Report. Functional Foods: Opportunities and challenges. Institute of Food Technologists, Chicago. http://members.ift.org/IFT/Research/IFTExpertReports/functionalfoods_report.htm.
Kauwell, G.P.A. 2005. Emerging concepts in nutrigenomics: A preview of what is to come. Nutr. Clin. Pract. 20: 75-87.
Kliewer, S.A., Xu, H.E., Lanbert, M.H., and Wilson, T.M. 2001. Peroxisome proliferator-activated receptors: from genes to physiology. Recent Prog. Horm. Res. 56: 239-263.
Marshall, E. 2004. Getting the noise out of gene arrays. Science 306: 630-631.
Matsui, N., Ito, R., Nishimura, E., Yoshikawa, M., Kato, M., Kamei, M., Shibata, H., Matsumoto, I., Abe, K, and Hashizume, S. 2005. Ingested cocoa can prevent high-fat diet-induced obesity by regulating the expression of genes for fatty acid metabolism. Nutrition 21: 594-601.
Müller, M. and Kersten, S. 2003. Nutrigenomics: Goals and strategies. Nature Rev. 4: 315-322.
Ordovas, J.M., Corella, D., Cupples, L.A., Demissie, S., Kelleher, A., Coltell, O., Wilson, P.W., Schaefer, E.J., and Tucker, K. 2002. Polyunsaturated fatty acids modulate the eff ects of the APOA1-G-A polymorphism on HDL-cholesterol concentrations in a sex-specifi c manner: The Framingham study. Am. J. Clin. Nutr. 75: 38-46.
Ouamrane, L., Larrieu, G., Gauthier, B., and Pineau, T. 2003. RXR activators molecular signaling: involvement of PPARa-dependent pathway in the liver and kidney, evidence for an alternative pathway in the heart. Brit. J. Pharmacol. 138: 845-854.
Pegorier, J.P., Le May, C., and Girard, J. 2004. Control of gene expression by fatty acids. J. Nutr. 134: 2444S-2449S.
Subbaramaiah, K., Bulic, P., Lin, Y., Dannenberg, A.J., and Pasco, D.S. 2001. Development and use of a gene promoter-based screen to identify novel inhibitors of cyclooxygenase-2 transcription. J. Biomolec. Screen. 6: 101-110.
Zeng, H., Davis, C.D., and Finley, J.W. 2003. Eff ect of selenium-enriched broccoli diet on diff erential gene expression in min mouse liver. J. Nutr. Biochem. 14: 227-231.
Zhu, M., Cabo, R., Anson, R.M., Ingram, D.K., and Lane, M.A. 2005. Caloric restriction modulates insulin receptor signaling in liver and skeletal muscle of rat. Nutrition 21: 378-388.
