Rapid Pathogen Screening Tools For Food Safety
Novel spectral- and mammalian cell-based techniques may offer faster detection of multiple and unknown pathogens in pre-harvest and post-harvest products.
Frequent food pathogen outbreaks and product recalls exert a huge economic burden on the United Sates economy, accounting for billions of dollars of losses each year. Application of broadband rapid pathogen screening tools to both pre-harvest and post-harvest products may be an effective strategy to produce safer foods. Spectral- or mammalian cell-based methods are ideal for high-throughput screening of microbes or toxins. Spectral-based light scattering sensors generate scatter signature of colonies of bacteria for detection, while cell-based biosensors measure interaction of active pathogens or toxins with mammalian cells.
Why Rapid Broadband Screening?
In general, most ready-to-eat food products are expected to be free of pathogens and a majority of raw or unprocessed products, with the exception of a few (e.g., Campylobacter in poultry), are generally free of known pathogens. Under such conditions, it does not make good economic sense for producers to test each batch of product for a specific pathogen, given the fact that each pathogen-specific kit is expensive and the procedure is lengthy. Thus, it may be undesirable for use with products that have limited shelf life. Moreover, a product may be negative for one pathogen but may be positive for another for which it has not been tested.
Therefore, cost-effective rapid high-through-put prescreening tools (Kim et al., 2008; Bhunia et al., 2010; Lee et al., 2010) could be used to sort out products that are free of harmful agents and subsequently released for consumption, while the suspect products can be tested with more specific analytical kits.
Application of a broadband pathogen screening tool is also sensible when the nature of the contaminants is not well defined, especially in pre-harvest food. Food production and supply chains are diverse and these may influence the nature of contaminants present. For example, operations for dairy or livestock production is different from fruits and vegetables production and each has unique on-farm practices, harvesting strategies, processing sequences, storage and handling practices, and distribution networks. Pathogen contamination can occur at any point during production, and the type and the nature of the pathogens may also vary. Implementing prescreening tools strategically throughout the production chain would enable producers to take immediate action to mitigate the problem, rather than waiting to do the testing at the end.
Furthermore, the CDC’s updated foodborne pathogen statistics serve as a continual reminder that of the 47.8 million cases of foodborne illnesses annually, an alarmingly high 38.4 million are caused by unspecified agents (Scallan et al., 2011). That means current methods are ineffective in monitoring the presence of some unknown biohazards. Moreover, with the escalation of potential bioterrorism through food and water, deployment of broadband rapid detection technology to serve as a first line of defense makes good sense.
A majority of commercially available assay kits detect only one pathogen at a time. For multi-pathogen analysis, one has to use many assay kits. This adds to the cost and labor of the testing. Therefore, recent research is geared towards developing an assay kit that can simultaneously detect the most common multiple pathogens, such as Salmonella, Shiga toxin-producing E. coli, Campylobacter, and Listeria. Eventually, this approach needs to be broadened to include other pathogens and toxins. However, hurdles related to uniform enrichment of all target organisms in a single medium, culture competition, and cross-reactions still need to be worked out.
--- PAGE BREAK ---
Recent Advances in Pathogen Testing
Traditionally, bacterial detection and identification are based on growth, biochemical properties, serology, and genome sequences. The serological approaches continue to evolve and antibodies are being used on dipstick assay, on paramagnetic beads, and on biosensor platforms for screening (Banada and Bhunia, 2008; Bryne et al., 2009). Bacteriophage proteins or pathogen-specific host cell receptor proteins are also being used as alternate molecules to antibodies. In nucleic acid-based methods, unique primers/probes are generated from the genome sequence to detect, identify, and quantify the pathogen by using polymerase chain reaction (PCR) in uniplex or multiplex formats (Levin, 2009). Furthermore, increasing genomic and proteomic information has enabled researchers to develop nucleic acid or protein microarrays for multi-pathogen testing. However, these methods require improved sample preparation strategies to eliminate food-derived inhibitors.
Biosensor methods—including optical, mass-based, electrochemical sensors—have been developed for pathogen detection (Bhunia, 2008; Kim et al., 2008; Koo et al., 2009; Privett et al., 2010; Bhunia et al., 2010). However, only a few show potential for high-throughput, real-time screening for multiple pathogens. The majority of sensors require an antibody or nucleic acid as a probe to provide specificity or fluorophores for an amplified signal. Specificity provided by a probe for a known target is desirable, but it precludes detection of emerging or genetically modified pathogens if present in the sample. Surface plasmon resonance, fiber optic, quartzcrystal microbalance, and impedance-based sensors are examples that require probes for interrogation of analytes (Banada and Bhunia, 2008; Bhunia et al., 2010).
Spectral-based methods, on the other hand, discriminate microbes based on the shape, molecular structure, color, and cellular organization. Sensors that include light scattering, Fourier transformed infrared spectroscopy, and Raman can detect microbes directly without any labeling reagents or probes. These sensors utilize lasers to generate characteristic absorption spectra of the sample or the wavelength frequency shift to accurately classify a species, serovar, or strain (Yu and Irudayaraj, 2006; Bhunia, 2008; Golightly et al., 2009; Bhunia et al., 2010).
Following are descriptions of the application of a spectral-based, novel light-scattering sensor and mammalian cell-based sensor as potential high-throughput pathogen screening tools.
Light-Scattering Sensor
The laser-light-scattering sensor, BARDOT (BActerial Rapid Detection using Optical scattering Technology), detects and identifies bacterial colonies as they grow on a Petri dish (Figure 1). Thus, this method can be easily integrated with routinely used culture-based methods (Banada et al., 2009). Laser scattering involves three steps: sample preparation, forward scatter measurement, and image processing.
The sample-preparation strategy is similar to what is recommended by the official USDA or FDA methods. However, the enrichment time may vary. Food rinsate or homogenate in appropriate enrichment broths are incubated for variable time periods (0–12 hr). Although a lengthy enrichment step is considered a bottleneck for most rapid tests, it is necessary even for a brief period to allow revival of injured or stressed bacteria, to increase target population, and to reduce background microflora.
--- PAGE BREAK ---
Since BARDOT can find a few colonies in the presence of other non-target bacteria on a plate, the normally used full-term (usually overnight) enrichment is not necessary. Enriched samples are then dispersed on nonselective or various selective agar plates, depending on the pathogens of interest for colony growth. Immunomagnetic separation techniques can also be used to concentrate target bacteria from the enriched samples. Filtrates from water, milk, or beverages can be directly applied to agar plates for colony growth. Likewise, clinical specimens or bodily fluids can be applied to plates for rapid diagnosis.
A Petri dish containing colonies (~1 mm in diameter) is placed under laser and the forward-scattering signatures of colonies are acquired via a charge-coupled device (CCD) imaging sensor (Bae et al., 2007). The image-processing software allows quantitative analysis followed by identification of bacterial type based on Zernike polynomials (Bayraktar et al., 2006). Recently, more advanced software based on a support vector machine allowed improved classification and identification of test organisms (Rajwa et al., 2010).

Initial studies have shown that BARDOT can successfully differentiate and classify foodborne bacteria from five major genera: Listeria, Salmonella, Escherichia, Staphylococcus, and Vibrio, representing 56 species and 95 strains/serovars (Banada et al., 2009). BARDOT is also able to differentiate various species of Listeria and Vibrio with greater than 94–99% accuracy. Recent unpublished work also shows evidence that it can discriminate at least the top 10 serovars of Salmonella. The ability of BARDOT (Figure 1) to detect and identify the aforementioned pathogens from inoculated ready-to-eat meats, red meat, chicken, leafy greens, eggs, and seafood samples has been demonstrated (Banada et al., 2009).
Mammalian Cell-based Functional Biosensor
Pathogens and toxins induce damage to the host cells during infection. Mammalian cells, naïve or engineered, have the ability to respond to pathogens in a physiologically relevant manner and can accurately estimate the pathogen’s associated risk (Banerjee and Bhunia, 2009; Bhunia, 2010). Thus, the use of mammalian cells makes it a functional biosensor.
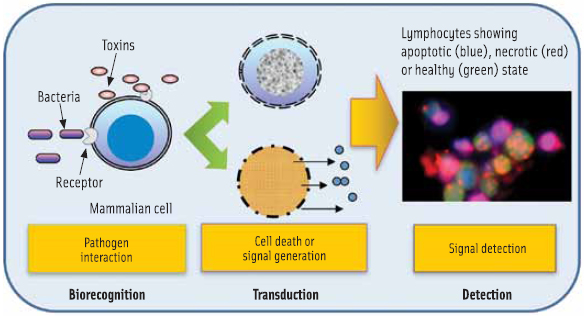
Traditional detection methods may not be suitable for addressing the changing scenario of food safety and biosecurity needs, such as detection of emerging pathogens, genetically modified microbes, or unknown or little known biohazards. A mammalian cellbased biosensor (CBBS) has the potential to fulfill such needs because this assay, unlike others, interprets pathogenic potential of viable microorganisms or active toxins rapidly (Figure 2). Also, there is no requirement for specific molecular probes for CBBS to be effective. Depending on the pathogen and the mammalian cells used, the cellular response could vary, which are characterized as altered cell shape, loss of surface molecules, inhibition of protein synthesis, cell death via apoptosis or necrosis, and cell lyses. These properties can be used as indicators to reveal the nature of toxicants such as those broadly classified as membrane pore-forming, diarrheagenic, superantigens, neurotoxin, or protein synthesis inhibitory toxin. However, the most effective way to exploit the potential of CBBS is to use appropriate cell types in an array format (Figure 3) that are most sensitive to a group of pathogens/toxins. The use of a mouse lymphocyte cell line was able to detect a broad range of toxins from Listeria monocytogenes, Bacillus spp., Staphylococcus aureus, Clostridium perfringens, and sea anemone or Stoichactis helianthus at nanogram quantities within 1–3 hr (Banerjee and Bhunia, 2010).
--- PAGE BREAK ---
Alternatively, the mammalian cells can be engineered to express specific surface receptors to interact with desired analytes. This interaction will initiate a downstream signaling event, leading to a cascade of reactions and resulting in a signal that will facilitate the identification or classification of the analyte.
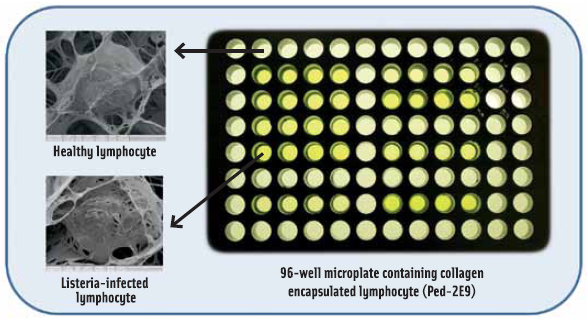
As part of the CBBS development, progress in culturing cells on biosensor platforms in three dimensional (3D) scaffolds mimicking cellular organization in organs and tissues is receiving increasing enthusiasm. Cells in 3D scaffold maintain a predetermined porosity to permit selective transport of analytes to the cells, while keeping coarse food particles at bay. Importantly, it produces more realistic in vivo type response. The use of a lymphocyte cell line in 3D scaffold on a 96-well micro plate (Figure 3) demonstrates that high-throughput screening of toxins at nanogram quantities and bacteria at 100–1,000 cells/g/ml from foods and beverages is possible (Banerjee and Bhunia, 2010).
The advantages of CBBS include rapid results, possibly in a few minutes to hours; positive response from a broad range of viable/active bioanalytes, unknown or genetically altered microbes; and improved sensitivity. Broad pathogen-specific response would also benefit the first responders to take necessary actions in the event of an outbreak.
Future Perspectives
Implementation of a food product prescreening strategy during preharvest and post-harvest production has the potential to reduce foodborne pathogen-related illnesses and product recalls. Several important criteria should be considered while developing prescreening tools. These should provide results in real-time or near real-time, be broad pathogens/toxins specific, have potential for application on-line or in-line, and be cost effective.
The laser-based light-scattering technology, BARDOT, allows real-time identification of bacterial colonies as they grow on a Petri dish. With an initial investment of the instrument, the cost for sample analysis would be minimal since it requires only routinely used microbiological media and reagents. Unlike other methods, BARDOT does not destroy the sample, keeping it intact for other tests if necessary. BARDOT requires bacterial growth; total assay time starting with the food sample may take 12–30 hr, depending on the growth rate. Recent work, however, demonstrated BARDOT’s ability to detect microcolony (100–200 micron diameter) could shorten the total detection time to 8 hr or less (Bae et al., 2010). A fully automated BARDOT system (Figure 1) with a built-in incubator could screen plates without user intervention and transfer data via the Internet for a decision to be rendered about a product “release” or “hold”.
CBBS responds to broad range of active biohazards but not to the inactive toxins or dead or harmless microbes, making it an ideal tool for high-throughput screening. Also, it would be an attractive tool for the first responders or food inspectors to initiate immediate counter measures in the event of an outbreak with bio-threat agents administered intentionally or inadvertently. CBBS could also be made highly specific by using appropriate naïve or engineered cell lines. However, the challenge is to prolong the CBBS life outside the laboratory, since the cells have stringent growth requirements.
Arun K. Bhunia, a Professional member of IFT, is Professor of Food Microbiology, Molecular Food Microbiology Laboratory, Center for Food Safety Engineering, Dept. of Food Science, Purdue University, West Lafayette, IN 47907 ( [email protected] ).
Acknowledgments
The research is supported by funds from USDA (1935-42000-035), NIH (1R56A1089511-01), and the Center for Food Safety Engineering at Purdue University.
References
Bae, E., Bai, N., Aroonnual, A., Bhunia, A.K., and Hirleman, E.D. 2010. Label-free identification of bacterial microlonies via elastic scattering. Biotechnol. Bioeng. Published online Nov. 2010 (DOI 10.1002/bit.22980).
Bae, E., Banada, P.P., Huff, K., Bhunia, A.K., Robinson, J.P., and Hirleman, E.D. 2007. Biophysical modeling of forward scattering from bacterial colonies using scalar diffraction theory. Appl. Opt. 46: 3639-3648.
Banada, P.P. and Bhunia, A.K. 2008. Antibodies and immunoassays for detection of bacterial pathogens. In: Zourob, M., Elwary, S., and Turner, A. (ed) Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems, Cambridge University, Manchester, UK.
Banada, P.P., Huff, K., Bae, E., Rajwa, B., Aroonnual, A., Bayraktar, B., Adil, A., Robinson, J.P., Hirleman, E.D., and Bhunia, A.K. 2009. Labelfree detection of multiple bacterial pathogens using light-scattering sensor. Biosens. Bioelectron. 24: 1685-1692.
Banerjee, P. and Bhunia, A.K. 2009. Mammalian cell-based biosensors for pathogens and toxins. Trends Biotechnol. 27: 179-188.
Banerjee, P. and Bhunia, A.K. 2010. Cell-based biosensor for rapid screening of pathogens and toxins. Biosens. Bioelectron. 26: 99-106.
Bayraktar, B., Banada, P.P., Hirleman, E.D., Bhunia, A.K., Robinson, J.P., and Rajwa, B. 2006. Feature extraction from light-scatter patterns of Listeria colonies for identification and classification. J. Biomed. Opt. 11: 034006.
Bhunia, A.K. 2008. Biosensors and bio-based methods for the separation and detection of foodborne pathogens. Adv. Food Nutr. Res. 54: 1-44.
Bhunia, A.K. 2010. Mammalian cell-based sensors: a rapid screening tool for pathogens and toxins in food. Food Eng. Ingredients. Oct. issue: 15-17.
Bhunia, A.K., Nanduri, V., Bae, E., and Hirleman, E.D. 2010. Biosensors, Foodborne Pathogen Detection. In: Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology. Flickinger, M.C. (Ed), John Wiley & Sons, Inc., Hoboken, NJ.
Byrne, B., Stack, E., Gilmartin, N., and Kennedy, R.O. 2009. Antibody-based sensors: Principles, problems and potential for detection of pathogens and associated toxins. Sensors 9: 4407-4445.
Golightly, R.S., Doering, W.E., and Natan, M.J. 2009. Surface-enhanced Raman spectroscopy and homeland security: A perfect match? ACS Nano 3: 2859-2869.
Kim, M.S., Chen, Y.R., Cho, B.K., Lefcourt, A.M., Chao, K.L. and Yang, C.C. 2008. Online hyperspectral line-scan fluorescence imaging for safety inspection of apples. In: Hewett, E.W., Lurie, S., and Wunsche, J.N. (ed) Proceedings of the International Symposium on the Role of Postharvest Technology in the Globalization of Horticulture.
Lee, M.R.F., Theobald, V.J., Ougham, H.J., Dahl, A.V., Lundby, F., Scollan, N.D., and Wold, J.P. 2010. Natural faecal fluorophores and the potential of chlorophyll based markers to optimise fluorescence as a real-time solution for the detection of faecal contamination on carcasses. Meat Sci. 86: 966-975.
Levin, R.E. 2009. Rapid Detection and Characterization of Foodborne Pathogens by Molecular Techniques. CRC Press. Boca Raton, FL.
Koo, O.K., Liu, Y.S., Shuaib, S., Bhattacharya, S., Ladisch, M.R., Bashir, R., and Bhunia, A.K. 2009. Targeted capture of pathogenic bacteria using a mammalian cell receptor coupled with dielectrophoresis on a biochip. Anal. Chem. 81: 3094-3101.
Privett, B.J., Shin, J.H., and Schoenfisch, M.H. 2010. Electrochemical Sensors. Anal. Chem. 82: 4723-4741.
Rajwa, B., Dundar, M.M., Akova, F., Bettasso, A., Patsekin, V., Hirleman, E.D., Bhunia, A.K., and Robinson, J.P. 2010. Discovering the unknown: Detection of emerging pathogens using a label-free light-scattering system, Cytometry 77A, 1103-1112.
Scallan, E., Griffin, P.M., Angulo, F.J., Tauxe, R.V., and Hoekstra, R.M. 2011. Foodborne illness acquired in the United States-unspecified agents. Emerg. Infect. Dis. (DOI:10.3201/eid1701.P21101).
Yu, C. and Irudayaraj, J. 2006. Identification of pathogenic bacteria in mixed cultures by FTIR spectroscopy. Transact. ASABE 49: 1623-1632.
