Next-Generation Sequencing Methods Revolutionize Food Microbiology
Application of whole genome sequencing technologies to food safety and food microbiology will dramatically change how we detect, subtype, and characterize foodborne pathogens, spoilage organisms, and starter cultures.
The first organism to have its entire genome sequence published was the RNA bacteriophage MS2 (in 1976); the first genome sequence from a DNA bacteriophage (Φ174) was published in 1977. Later that year, Sanger and colleagues published an article detailing the chain-termination sequencing technique, often called the Sanger sequencing method. For more than 20 years, the Sanger sequencing method was the only sequencing method available. The first bacterial genome sequenced by this method, Haemophilus influenzae, was published in 1995.
The first human genome was also sequenced by the Sanger method; it took more than 10 years to complete the sequencing of approximately 3 billion bases at a cost of approximately $3 billion. Recently, the average cost of sequencing a finished genome (to completion with annotation), using Sanger sequencing, has been estimated to be 10 cents/base pairs, so that the cost of sequencing of a 4 Mb (millions of base pairs) bacterial genome would be $400,000 (Service, 2006). In addition to the high cost, the Sanger sequencing method is impeded by low throughput, thus rendering the method limiting for large-scale population-based genomics projects and other practical applications of whole genome sequencing.
A 1996 publication entitled “Real-Time DNA Sequencing Using Detection of Pyrophosphate Release” represented a technological turning point that sparked the development of new rapid sequencing methods, which have revolutionized DNA sequencing and reinvigorated enthusiasm about practical applications of DNA sequencing in applied fields. Limitations of Sanger sequencing prompted the J. Craig Venter Foundation, X-prize Foundation, and National Institutes of Health to collectively challenge the biotechnology community to pioneer the next generation of sequencing chemistries and platforms in the early 2000s with the ultimate goal of generating a whole human genome sequence for <$1,000.
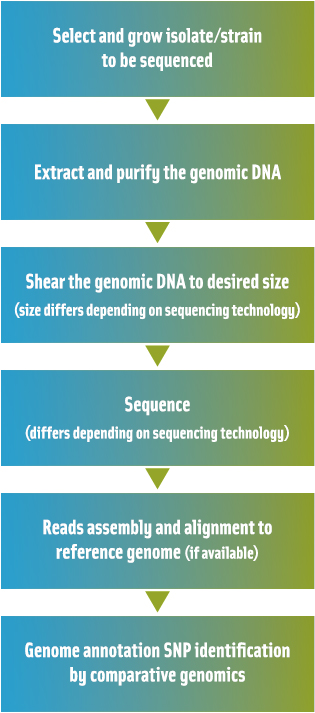 Several next-generation sequencing technologies have since been introduced, and these methods are significantly less costly, time consuming, and labor intensive as compared to the current gold standard Sanger method (Figure 1). In 2005, investigators from 454 Life Sciences (now partnered with Roche) and from the laboratory of George Church (Harvard Medical School) independently published two innovative, high-throughput sequencing-by-synthesis methods, which have been commercially available for a few years (i.e., the 454 Genome Analyzer and the Illumina [formerly Solexa Inc.] system). Since then, Applied Biosystems (now Life Technologies) released a sequencer called SOLiD (supported oligo ligation detection).
Several next-generation sequencing technologies have since been introduced, and these methods are significantly less costly, time consuming, and labor intensive as compared to the current gold standard Sanger method (Figure 1). In 2005, investigators from 454 Life Sciences (now partnered with Roche) and from the laboratory of George Church (Harvard Medical School) independently published two innovative, high-throughput sequencing-by-synthesis methods, which have been commercially available for a few years (i.e., the 454 Genome Analyzer and the Illumina [formerly Solexa Inc.] system). Since then, Applied Biosystems (now Life Technologies) released a sequencer called SOLiD (supported oligo ligation detection).
In December 2010, Life Technologies also acquired a new sequencing platform—the Ion Torrent Personal Genome Machine, which uses revolutionary semiconductor technology. While the pyrophosphate released during incorporation of dNTPs (deoxyribonucleotide triphosphate) forms the basis for the pyrosequencing methods, the Ion Torrent system detects the proton released during dNTP incorporation. The method works with natural dNTPs, has no enzyme cascading reactions, and detects protons directly using semiconductors. The instrument system is very cost-effective, as it has no optics and leverages years of investment in semiconductor chip manufacturing.
--- PAGE BREAK ---
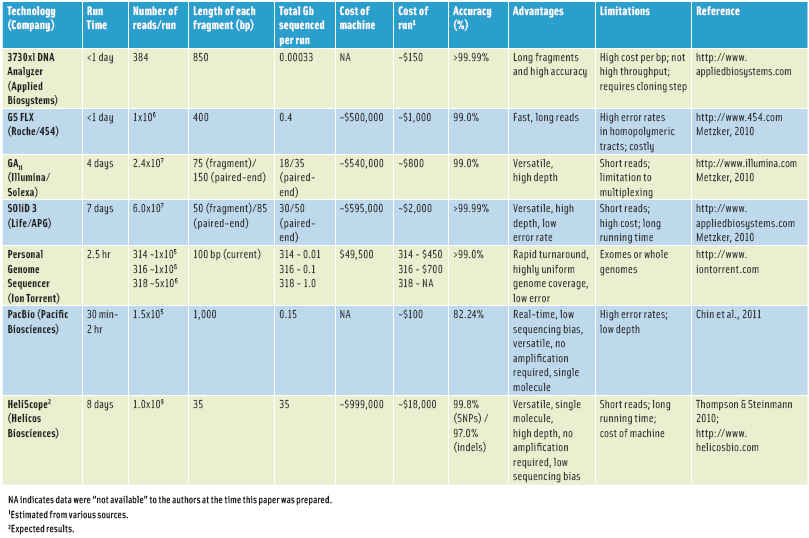 These four technologies are currently considered to be the so-called “second-generation” sequencing methods, which allow for completion of a draft human genome for <$10,000 (Table 1). A detailed review and comparison of these methods are beyond the scope of this article, but more in-depth reviews on next-generation sequencing technologies are available (e.g., Metzker, 2010).
These four technologies are currently considered to be the so-called “second-generation” sequencing methods, which allow for completion of a draft human genome for <$10,000 (Table 1). A detailed review and comparison of these methods are beyond the scope of this article, but more in-depth reviews on next-generation sequencing technologies are available (e.g., Metzker, 2010).
Currently, new “third-generation” sequencing systems, which are also known as single-molecule sequencing technologies, are in the process of commercialization, promising even greater improvements in throughput and cost reduction for whole genome sequencing. Examples of these systems include sequencing systems by Helicos and Pacific Biosciences (Table 1). It seems feasible that at least some of these systems will provide an avenue to meet the goal of sequencing a complete human genome for <$1,000. Since bacterial genomes are about 1,000-fold smaller than the human genome, these technologies will likely also allow for complete bacterial genome sequences at a cost similar to that of routine bacterial subtyping methods, including pulsed field gel electrophoresis (PFGE) and multilocus sequence typing (MLST).
Improved Detection Methods 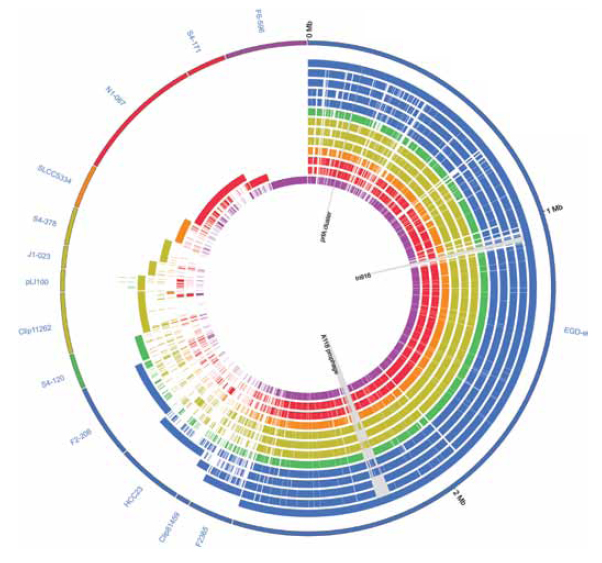 The ability to rapidly sequence the genomes of foodborne disease-causing organisms and closely related non-pathogenic organisms allows for rapid identification of genes and other markers that 1) are unique to the disease-causing organism and 2) are linked to or responsible for the ability of an organism to cause disease. Technically, this approach requires a combination of genome sequencing with advanced analyses known as comparative genomics (i.e., comparison of different genomes to identify common and unique features). This approach was, for example, used to identify unique features of the pathogen E. coli O157:H7 that differentiate it from its close, non-pathogenic relative E. coli O55:H7 (Zhou et al., 2010). More recently, these technologies have been used to identify unique features of Listeria spp. that cause human and animal disease (e.g., L. monocytogenes and L. ivanovii) and to differentiate disease-causing strains and species from non-pathogenic strains and species (Figure 2; den Bakker et al., 2010).
The ability to rapidly sequence the genomes of foodborne disease-causing organisms and closely related non-pathogenic organisms allows for rapid identification of genes and other markers that 1) are unique to the disease-causing organism and 2) are linked to or responsible for the ability of an organism to cause disease. Technically, this approach requires a combination of genome sequencing with advanced analyses known as comparative genomics (i.e., comparison of different genomes to identify common and unique features). This approach was, for example, used to identify unique features of the pathogen E. coli O157:H7 that differentiate it from its close, non-pathogenic relative E. coli O55:H7 (Zhou et al., 2010). More recently, these technologies have been used to identify unique features of Listeria spp. that cause human and animal disease (e.g., L. monocytogenes and L. ivanovii) and to differentiate disease-causing strains and species from non-pathogenic strains and species (Figure 2; den Bakker et al., 2010).
In addition, these comparative genomics approaches will also facilitate development of improved detection methods for shiga toxin-producing E. coli (STEC) belonging to serotypes other than O157 and will likely facilitate the development of serotype or strain specific assays for different Salmonella. In the not-too-distant future, it is likely that outbreak investigations will be able to rapidly deploy PCR-based assays that specifically detect an outbreak strain with PCR targets identified through rapid whole genome sequencing and comparative genomics.
Improved Subtyping
Molecular and DNA-based subtyping methods have had a considerable positive impact on food safety by providing tools that are now commonly used to detect foodborne disease outbreaks and that also can be used to identify contamination sources throughout the food chain. Commonly used subtyping methods for foodborne pathogens include PFGE, ribotyping, multilocus variable number tandem repeat analysis (MLVA) as well as a number of other PCR-based and DNA sequencing-based subtyping methods.
--- PAGE BREAK ---
Next-generation sequencing will likely have a considerable impact on molecular subtyping of food-associated organisms. Full bacterial genome sequences, including some that were generated with these methods, are already frequently used to develop subtyping methods for certain foodborne pathogens. For example, full genome sequence data have been critical for the development of MLVA methods for different pathogens that are difficult to differentiate by PFGE, such as certain Salmonella serotypes (e.g., Newport, Typhimurium, and Enteritidis), Bacillus anthracis, and Listeria monocytogenes isolates belonging to serotype 4b. In addition, full genome sequences have been critical for the development of MLST-based and single nucleotide polymorphism (SNP)-typing-based subtyping methods.
A first glimpse into the potential of genome sequencing as a subtyping tool for bacterial foodborne pathogens was provided by the approach used as part of the investigation into the 2001 B. anthracis bioterrorism incident in the U.S. Initial MLVA characterization of isolates from victims of the “letter-based” anthrax inhalation cases suggested a common source; an isolate from a cow in Texas in 1981 that was subsequently used as a laboratory control strain at Fort Detrick for research purposes. Since B. anthracis isolates show high nucleotide identity (>99%), comparative genome sequencing of a patient isolate and the lab control strain was performed by the Sanger method. Comparative genomics revealed only four mutations between the two strains (i.e., two SNPs and two indels), supporting the causal association between the lab strain and the outbreak strain (Read et al., 2002).
Since then, whole genome sequence data have been used in a retrospective study of a 1988 sporadic listeriosis case and a 2000 listeriosis outbreak that were both caused by the same L. monocytogenes PFGE type, which appears to have persisted for at least 12 years in the food production facility linked to both the 1988 case and the 2000 outbreak (Orsi et al., 2008). In this case, the full genome data showed that the 1988 human and food isolates were virtually identical to the 2000 human and food isolates, except for a single prophage (i.e., a phage integrated into the bacterial chromosome), which differed considerably between the 1988 and the 2000 isolates, suggesting a possible prophage replacement that occurred between 1988 and 2000. Full genome sequences were thus able to differentiate the 1988 and the 2000 strains, which was not possible with other subtyping methods (e.g., PFGE, ribotyping), and confirm a very close genetic relatedness (with differences that can easily emerge over short time periods on an evolutionary timescale (<12 years). This study thus confirmed the likely long-term persistence of L. monocytogenes in a food processing facility.
Consequently, next-generation approaches have been employed as full genome subtyping tools to investigate foodborne disease outbreaks, including the 2007 listeriosis outbreak in Canada (Gilmour et al., 2010). Although a majority of patient isolates associated with this outbreak shared identical PFGE patterns, one clinical isolate and several isolates from the environment of the food processing facility associated with the outbreak showed a closely related PFGE pattern that differed by two bands from the reference outbreak strain. Pyrosequencing of a human clinical isolate representing the reference outbreak PFGE type and a closely related human clinical isolate associated with the outbreak along with follow-up targeted sequencing revealed that the outbreak could be attributed to three closely related strains.
Most recently, whole genome sequencing was employed by the U.S. Food and Drug Administration as a subtyping tool to help investigate a salmonellosis outbreak, which was attributed to a common Salmonella Montevideo PFGE type. Comparative genomics further supported the causal link between salami formulated with contaminated pepper and the outbreak (Lienau et al., 2011). Another recent study used “third-generation” single-molecule real-time DNA sequencing to compare the genomes of five Vibrio cholerae strains involved in cholera episodes in Latin America in 1991, South Asia in 2002 and 2008, and the current outbreak in Haiti (Chin et al., 2011). Interestingly, the authors found that the V. cholerae strain causing the Haitian outbreak is more closely related to the strains isolated from South Asia in 2001 and 2008 than to the strain isolates from Latin America in 1991, suggesting that the strain causing the Haitian outbreak was introduced in Haiti by human activity.
--- PAGE BREAK ---
Understanding Food-Associated Microbes
The ability to rapidly generate full genome sequence data for a number and, often large, number of strains of a given organism provides a tremendous opportunity to gain a better understanding of the biology of food-associated microorganisms. For example, a recent study (den Bakker et al., 2010) used newly generated and assembled genome sequences for a number of Listeria isolates representing either species not previously sequenced or strains with unusual phenotypes (e.g., hemolytic L. innocua) to provide considerable new knowledge about the biology and evolution of the genus Listeria. Comparative genomics analyses provided clear evidence that the current members of the genus Listeria appear to have evolved from a common ancestor that already carried most key genes required for virulence, through multiple independent losses of virulence genes, which resulted in a number of avirulent species (e.g., L. innocua). This study also defined a key set of genes that can be used to differentiate Listeria isolates that can cause disease in mammalian hosts from those that are restricted to a saprophytic lifestyle. Full genome sequence data can also be used to predict metabolic pathways and capabilities of a given organism or group of organisms (Henry et al., 2010). Findings described by Henry et al. (2010) not only provide insights into the products of bacteria that grow on foods, which will be of particular value for fermentation organisms, but could potentially also be used to predict the culture conditions a given organism or group of organisms can grow under. Ultimately, genome sequence data may thus facilitate the development of appropriate selective and differential media for the detection of a given target organism (or group of organisms).
Next-generation sequencing methods cannot only be used to rapidly sequence genomes, but also have been used to sequence the “transcriptome” (i.e., all RNA copies present) of bacteria grown under defined conditions, including one study that sequenced the transcriptome of a Listeria monocytogenes strain (Oliver et al., 2009). These uses of next-generation sequencing will allow in-depth studies on how bacteria respond to different environmental and food-associated conditions, which can, for example, be used to develop improved strategies to control and prevent microbial growth and survival in different foods. These applications of next-generation sequencing methods will likely rapidly replace microarrays, which are currently often used to characterize genome-wide transcriptional patterns and responses, as transcriptome sequencing overcomes a number of the shortcomings of microarrays.
Metagenomics
Next-generation sequencing methods allow for rapid large-scale population-based studies of food-associated and intestinal microbes. The ability of next-generation sequencing to generate more than 100 Mb of DNA sequence per run has facilitated the studies known as metagenomics. Metagenomics is a comprehensive approach to probe the whole microbial community (i.e., culturable and non-culturable microorganisms) present in virtually any sample (i.e., clinical and environmental samples). Before the emergence of high-throughput sequencing, metagenomic studies required the cloning and sequencing of short target fragments directly from a sample, usually ribosomal DNA loci, which made this approach very costly and time consuming. Nowadays, with the aid of high-throughput sequencing, the total microbial DNA can be extracted and sequenced directly from that sample without cloning.
Metagenomics has the potential to be applied to several areas of food safety and food microbiology, including the identification of 1) the causative agent of foodborne disease, 2) commensal microorganisms that may be associated with the presence or absence of a pathogen in a given niche, 3) shelf life-limiting spoilage organisms, and 4) microorganisms that are important for fermentation systems. For example, a recent study employed a metagenomics approach to identify Campylobacter jejuni as the causative agent for a foodborne illness case that was not diagnosable by conventional microbiological culture. Briefly, total microbial DNA was extracted from two fecal samples collected from a patient and comparative genomics analyses showed that C. jejuni DNA was present in the sample collected from the patient while he was experiencing clinical symptoms of foodborne illness but the organism was not detected from the fecal sample collected three months following clearance of the infection (Nakamura et al., 2008).
--- PAGE BREAK ---
Future Challenges
It is clear that next-generation sequencing approaches will have a considerable impact on many, if not all, areas in food microbiology and food safety. The challenges with these methods do not typically involve the actual generation of genome and transcriptome data, but rather lie in the ability to correctly handle, analyze, and interpret the extremely large, terabyte-scale, datasets that are generated by these methods. The ability to translate potentially exciting discoveries facilitated by next-generation sequencing to practical outcomes that improve food safety, quality, and sustainability thus requires “next-generation” food scientists and food microbiologists that not only understand food systems and food microbiology, but are also well-versed in bioinformatics and other quantitative disciplines.
Molecular Methods in Food Microbiology Symposium and Workshop
In addition to their research on molecular subtyping methods and molecular epidemiology, Martin Wiedmann (Cornell University) and Kendra Nightingale (Colorado State University) are also committed to providing training for current and future professionals in the food industry, government, and academic institutions in the areas of molecular detection and subtyping of food-associated microorganisms. As a result, Wiedmann and Nightingale developed and co-direct a workshop series on “Molecular Methods in Food Microbiology,” which is currently supported by a U.S. Dept. of Agriculture: Cooperative State Research, Education: National Integrated Food Safety Initiative grant.
To assure that this new workshop series meets current and future food safety training needs, Wiedmann and Nightingale partnered with scientists from the Silliker Food Science Center of Silliker Inc. and assembled an advisory board comprised of individuals from the food industry, regulatory agencies, and academia with particular expertise in applying molecular approaches to food microbiology. The Fourth Annual Molecular Methods in Food Microbiology Workshop and Symposium Series will be held at Colorado State University (Fort Collins, Colo.), June 27–July 1. It will specifically focus on use and application of DNA sequencing, including conventional Sanger and next-generation sequencing technologies, for molecular detection and subtyping of food-associated microorganisms. In addition, a half-day current topics session in this symposium will focus on detection and subsequent characterization of non-O157 shiga toxin-encoding Escherichia coli (STEC). The two-and-ahalf day hands-on laboratory session will focus on 16s rDNA sequence-based identification, multilocus sequence typing, design of a custom multiplex detection method for STEC or other targets of interest, and application of commercially available multiplex real-time PCR assays to detect foodborne pathogens. This course is appropriate for both individuals who have attended previous workshops and for people who have not attended the prior workshops.
For more details on the workshop schedule and to register online, please visit http://ansci.colostate.edu/department/Events/MMFM_2011/mmfm_2011.html.
Martin Wiedmann , Ph.D., a Professional Member of IFT, is Associate Professor, Dept. of Food Science, Cornell University, Ithaca, NY 14853 ([email protected]).
Renato H. Orsi is a Postdoctoral Research Associate, Dept. of Food Science, Cornell University, Ithaca, NY 14853 ([email protected]).
Manohar R. Furtado , Ph.D., is Vice President of Research and Development, Food and Environmental, Animal Health, Pharma Analytics and Molecular Diagnostics at Life Technologies ([email protected]).
Kendra K. Nightingale , Ph.D., a Member of IFT, is Assistant Professor, Dept. of Animal Sciences, Colorado State University, Fort Collins, CO 80523 ([email protected]).
Acknowledgements
Work on molecular subtyping in the author’s laboratories is supported by U.S. Dept. of Agriculture Special Research Grants (to MW) and a National Integrated Food Safety Initiative Grant (Grant No. 2008-51110-04333) of the Cooperative State Research, Education, and Extension Service, U.S. Dept. of Agriculture (to KKN and MW). Opinions, findings, conclusions, or recommendations expressed in this article are those of the authors and do not necessarily reflect the U.S.D.A.
References
Chin, C.S., Sorenson, J., Harris, J.B., Robins, W.P., Charles, R.C., Jean-Charles, R.R., Bullard, J., Webster, D.R., Kasarskis, A., Peluso, P., Paxinos, E.E., Yamaichi, Y., Calderwood, S.B., Mekalanos, J.J., Schadt, E.E., and Waldor, M.K. 2011. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364: 33-42.
Den Bakker, H.C., Cummings, C.A., Ferreira, V., Vatta, P., Orsi, R.H., Degoricija, L., Petrauskene, O., Furtado, M.R, and Wiedmann, M. 2010. Comparative genomics of the bacterial genus Listeria: Genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics. 2:688.
Gilmour, M.W., Graham, M., Van Domseiaar, G., Tyler, S., Kent, H., Trout-Yakel, K.M., Larios, O., Allen, V., Lee, B., and Nadon, C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics. 11: 120.
Henry, C.S., DeJongh, M., et al. 2010. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotech 28: 977-982.
Lienau, E.K., Strain, E., Wang, C., Zheng, J., Ottesen, A.R., Keys, C.E., Hammack, T.S., Musser, S.M., Brown, E.W., Allard, M.W., Cao, G., Meng, J., and Stones, R. 2001. N. Engl. J. Med. 364: 981-982l.
Metzker, M.L. 2010. Sequencing technologies – the next generation. Nat. Rev. Genet. 11: 31-46.
Nakamura S., Maeda, N., Miron, I.M., Yoh, M., Izutsu, K., Kataoka, C., Honda, T., Yasunaga, T., Nakaya, T., Kawai, J., Hayashizaki, Y. Tosshihiro, H., and Lida, T. 2008. Metagenomic diagnosis of bacterial infections. Emerg. Infect. Dis. 14: 1784-1786.
Oliver, H.F., Orsi, R.H., Ponnala, L., Keich, U., Wang, W., Sun, Q., Cartinhour, S.W., Filiatrault, M.J., Wiedmann, M., and Boor, K.J. 2009. Deep RNA sequencing of Listeria monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics. 10: 641.
Orsi, R.H., Borowsky, M., Lauer, P., Young, S.K., Nusbaum, C., Galagan, J.E., Birren, B.W., Ivy, R.A., Sun, Q., Graves, L.M., Swaminathan, B., and Wiedmann, M. 2008. Shortterm genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics. 9: 539.
Read, T.D., Salzberg, S.L., Pop, M., Shumway, M., Umayam, L., Jiang, L., Holtzapple, E., Busch, J.D., Smith, K.L., Schupp, J.M, Solomon, D., Keim, P., and Fraser, C.M. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science. 296: 2028-2033.
Service, R.F. 2006. Gene sequencing. The race for the $1,000 genome. Science. 311: 1544-1546.
Thompson, J.F. and Steinmann, K.E. 2010. Single molecular sequencing with a HeliScope genetic analysis system. Curr. Prot. Mol. Biol. Chapter 7. Unit 7.10.
Zhou, Z., Li, X, Liu, B., Beutin, L., Xu, J., Ren, Y., Feng, L., Lan, R., Reeves, P.R., and Wang, L. 2010. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLoS One. 5: e8700.
