Deepening Analysis of Foodborne Bacteria
FOOD SAFETY AND QUALITY
Food producers, processors, and distributors want increasingly rapid microbial results to mesh with their just-in-time supply chains.
To meet this demand, companies are developing and marketing simple, user-friendly tests that can be performed at the processing plant or in the field by relatively untrained personnel. Primed and ready to replace culture and molecular screening methods, biosensor-based analyses using specific biomarkers such as nucleic acids, proteins, antigens, and metabolic products are the third wave of microbial detection systems. These kits and biosensor formats target the food industry’s goal of quickly and inexpensively monitoring safety and quality, then moving the product out the door.
But before the food industry slashes testing budgets, stakeholders should consider a paradigm shift in food safety and agricultural research: Microbial testing should go not only faster but deeper.
Deep Means Sequencing
Culture methods, unfortunately, have significant drawbacks for identifying all the individual species in a microbiological community. Not all microbes will grow in a lab environment, and some damaged or stressed organisms may be viable but not culturable. Colony counting methods cannot reliably enumerate or identify microbial cells such as spores, which tend to aggregate and clump when plated. Different food matrices are challenging for culture methods, and spices, herbs, chocolate, and vitamin and mineral premixes are known to inhibit microbial growth and confound the reading of culture films.
Deep analysis by sequencing circumvents cultural limitations by going directly to nucleic acids such as DNA and RNA for identification. Discovering and cataloging the diversity of pathogenic serovars and strains, microbiomes, biofilms, and other microbial communities may help prevent recalls, mitigate outbreaks, assist in the discovery of antimicrobial-resistant microbes, and point to contaminants, formulation, or environmental changes (Beck et al. 2021, Thompson et al. 2018).
Sequencing also has become more affordable and accessible. High-throughput sequencing of total DNA and total RNA is leaving the realm of academics and moving into use in industry. In addition, novel technologies such as CRISPR-SeroSeq provide deeper information (Thompson et al. 2018).
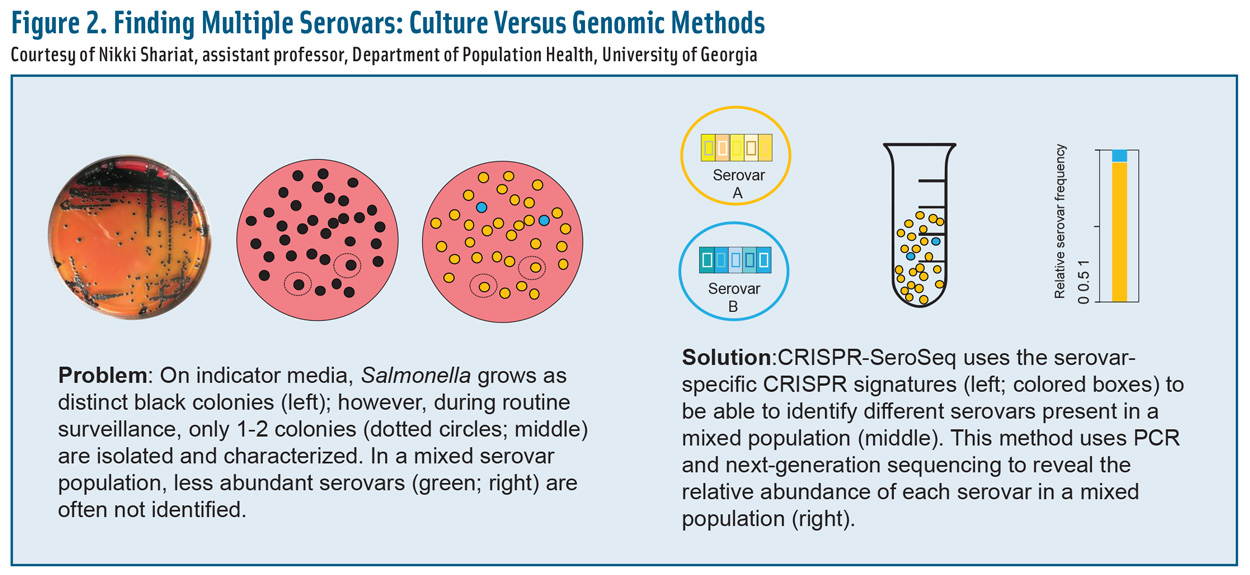
Culture-Based Salmonella Serotyping
Most serotyping has been limited to culture-based approaches that detect only the most abundant or culturable serovars or strains. One recent study, however, found that 91% of samples taken from poultry houses and processing facilities harbor more than one serovar (Thompson et al. 2018). In fact, up to nine serovars have been found on a single broiler carcass (Rasamsetti et al. 2021).
Statistically speaking, six colonies would need to be picked and serotyped to ensure a 95% probability of identifying two serovars (Cason Jr. et al. 2011), but that probability is valid only if both serovars are in equal numbers in the sample. If one of the serovars is outnumbered 10:1 by another, it would take at least 32 colonies for the minority serovar to be discovered (Cason Jr. et al. 2011).
Multiplex Salmonella Serotyping
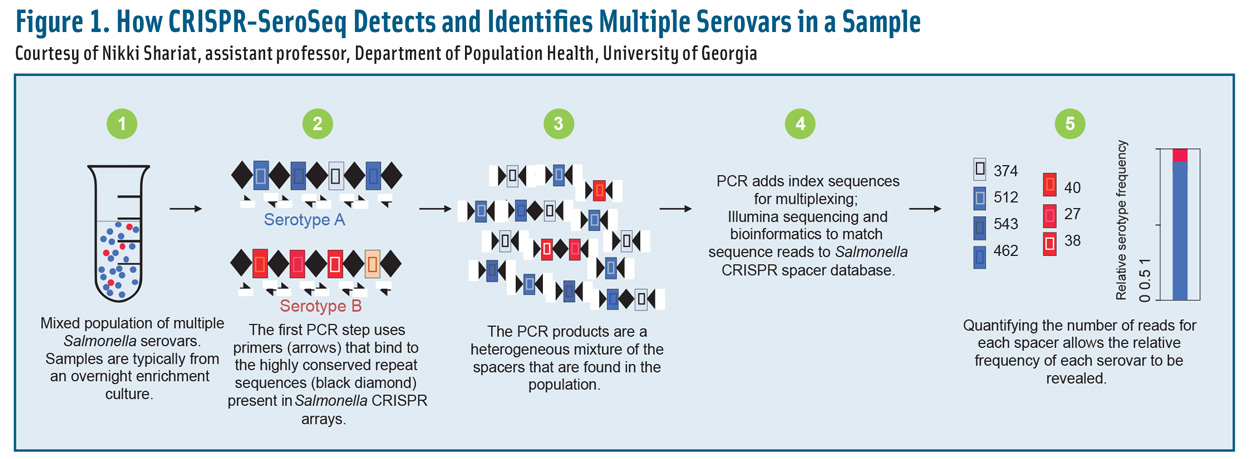
The discovery of clustered regularly interspaced short palindromic repeat (CRISPR)-Cas systems has revolutionized DNA editing through precise targeting, cutting, and splicing of genes of interest in living cells (Barrangou and Horvath 2017). A prokaryotic adaptive immune system, CRISPR-Cas is found in approximately 45% of sequenced bacterial genomes (Grissa et al. 2007). CRISPR arrays contain highly variable spacer sequences surrounded by relatively stable unvarying repeat sequences. The spacer regions are short portions of foreign DNA that were acquired when the Salmonella CRISPR-Cas system was actively protecting the cell from invading bacteriophages (Shariat et al. 2015).
Salmonella species have two arrays, named CRISPR1 and CRISPR2, which have been shown to have distinct serovar-specific spacer regions (Fabre et al. 2012, Touchon and Rocha 2010). These regions can be amplified by universal PCR primers based on the unvarying repeat sequences. After sequencing, the DNA information can be compared with a growing database that identifies the serovar (Thompson et al. 2018). CRISPR-SeroSeq is the name of this novel molecular tool that can investigate the depth of serovar diversity down to 0.01% of the sample (Thompson et al. 2018).
Salmonella CRISPR arrays can be used because researchers noticed that these bacteria are no longer adapting to phage attack by acquiring new spacer regions (Shariat et al. 2015). The genomic sequences in this region of interest are stable, but crucially, they differ among different serovars.
“Our goal is that the use of CRISPR-SeroSeq will buy us more time to mitigate a potential foodborne Salmonella outbreak,” says Nikki Shariat, a scientist at University of Georgia’s Poultry Diagnostic and Research Center and lead developer of the assay. “By monitoring production and processing facilities, we can detect serovars that are increasing long before we are able to routinely pull the colonies off a plate. We hope this will allow us to predict an outbreak before it becomes a problem.”
Bacterial communities are complex and constantly evolving due to changes in the environment, available growth matrices, and factors such as water activity and pH. Over time, certain microbes tend to predominate and become more abundant than others. Figure 1 illustrates how molecular deep dives into serovar diversity can identify changes in serovar abundance more rapidly than culture-based methods can. This predictive system may also be helpful in providing an early warning when a highly virulent or antibiotic-resistant serovar is on the rise.
For example, a CRISPR-SeroSeq study looking at cattle feces samples found a minority serovar, Salmonella Reading, that was tetracycline resistant (Siceloff et al. 2021). Fortunately, the abundant serovars in the cattle feces were susceptible to the antibiotic (Siceloff et al. 2021). However, this knowledge may be used to mitigate the increase and predominance of the tetracycline-resistant strain.
CRISPR-SeroSeq is also more cost-effective than metagenomics, a less biased sequencing method, because CRISPR-SeroSeq can multiplex several samples on a single sequencing run (Thompson et al. 2018). This significantly lowers sequencing costs and makes it more attractive to monitor Salmonella serovar diversity over time.
In an effort to expand the CRISPR sequence database, Shariat’s lab continues to analyze CRISPR sequences in additional serovars and add them to the database. Since the initial publication of the method, the database has increased from 109 to 130 serovars, according to Shariat.
Microbiome Shifts
Medical research has shown that the composition of the gut microbiome is indicative of a change in an individual’s diet, environment, or health. So food scientists reasoned that a shift in the food microbiome could be used as an indicator of contaminants or changes in the environment (Beck et al. 2021).
They tested this hypothesis by sequencing the total RNA of 31 high protein powder (HPP) samples derived from poultry meal. Using a novel microbiome analysis pipeline, they identified 119 microbial genera and found that Bacteroides, Clostridium, Lactococcus, Aeromonas, and Citrobacter were the most abundant species. Shifts in the microbial community occurred when ingredient compositions differed—specifically during matrix contamination, when pork and beef unexpectedly were found in the HPP. These matrix-contaminated HPP samples had increased relative abundances of Lactococcus, Lactobacillus, and Streptococcus.
“It is through shifts in the microbiome that potential pathogen outbreaks can be identified, whether as a result of environmental shifts or intentional adulteration,” says Bala Ganesan, metagenomics and bioinformatics principal scientist at the Mars Global Food Safety Center. “Crucially, this approach increases visibility to elements that could pose a food safety threat and gives advance insights needed to take action.”
Sequenced DNA or RNA is a molecular test and not necessarily indicative of viable microorganisms, so quantification of Salmonella in the HPP samples determined by total RNA sequencing data and culture-based assays was also compared (Beck et al. 2021). A correlation between molecular and culture-based data was not apparent (Beck et al. 2021). Nevertheless, total RNA sequencing was considered a useful tool for cataloging microbiome membership in a sample, and it could be employed to detect food adulteration, food fraud, and mislabeling.
Harnessing Technology for the Future
As powerful technologies such as sequencing become affordable for routine use, more producers will take advantage of the deep and predictive capabilities of these molecular detection methods. Partnered with bioinformatics, DNA and RNA sequencing can provide more complete microbial profiles and coherent histories. These data are not only predictive but diagnostic, allowing the producer to uncover hidden problems.
In a recent profile in Food Safety News (Koger 2021), Drew McDonald, vice president of quality and food safety for Taylor Farms in Salinas, Calif., commented that in order to strive for true food safety improvement, industry members need to focus on data. “The real trick is that as that data is generated, you’re looking back at it, you’re using it to solve problems and probably more importantly, you’re using it to ask questions,” he said. “One of the expressions we have at Taylor Farms is that ‘we’re not necessarily getting better answers, but we’re getting really good at asking questions.’”
REFERENCES
Barrangou, R. and P. Horvath. 2017. “A Decade of Discovery: CRISPR Functions and Applications.” Nat. Microbiol. 2: 17092. doi: 10.1038/nmicrobiol.2017.92.
Beck, K. L., N. Haiminen, D. Chambliss, et al. 2021. “Monitoring the Microbiome for Food Safety and Quality Using Deep Shotgun Sequencing.” npj Sci. Food. 5(1): 3. doi: 10.1038/s41538-020-00083-y.
Cason Jr., J. A., N. A. Cox Jr., R. J. Buhr, et al. 2011. “Probability of Identifying Different Salmonella Serotypes in Poultry Samples.” Poultry Science Association Meeting Abstract.
Fabre, L., J. Zhang, G. Guigon, et al. 2012. “CRISPR Typing and Subtyping for Improved Laboratory Surveillance of Salmonella Infections. PLoS One. 7(5): e36995.
doi: 10.1371/journal.pone.0036995.
Grissa, I., G. Vergnaud, and C. Pourcel. 2007. “CRISPRFinder: A Web Tool to Identify Clustered Regularly Interspaced Short Palindromic Repeats.” Nucleic Acids Res. 35: W52–7. doi: 10.1093/nar/gkm360.
Koger, C. 2021. “Let Data Drive the Food Safety Process and Share Knowledge With the Industry.” Food Safety News, July 21.
Rasamsetti, S., M. Berrang, N. A. Cox, et al. 2021. “Selective Pre-Enrichment Method to Lessen Time Needed to Recover Salmonella From Commercial Poultry Processing Samples.” Food Microbiol. 99: 103818. doi: 10.1016/j.fm.2021.103818.
Shariat, N., R. E. Timme, J. B. Pettengill, et al. 2015. “Characterization and Evolution of Salmonella CRISPR-Cas Systems.” Microbiol. 2: 374–386. doi: 10.1099/mic.0.000005.
Siceloff, A. T., N. Ohta, K. N. Norman, et al. 2021. “Antimicrobial Resistance Hidden Within Multiserovar Salmonella Populations.” Microb. Ecol. 65(6). doi: 10.1128/AAC.00048-21.
Thompson, C. P., A. N. Doak, N. Amirani, et al. 2018. “High-Resolution Identification of Multiple Salmonella Serovars in a Single Sample by Using CRISPR-SeroSeq.” J. Appl. Environ. Microbiol. 21: e01859–18. doi: 10.1128/AEM.01859-18.
Touchon, M. and E. P. C. Rocha. 2010. “The Small, Slow and Specialized CRISPR and Anti-CRISPR of Escherichia and Salmonella.” PLoS One. 5: e11126. doi: 10.1371/journal.pone.0011126.