Enhancing Foods with Functional Genomics
Understanding how organisms express specific genes in different situations can lead to improvements in food production, processing, and safety.
Worldwide, genome sequencing projects focused on agriculturally related plants, animals, and microbes are progressing at a tremendously rapid pace. As these projects come to fruition, they will provide a wealth of information that will dramatically impact agriculture and food production.
The rate at which new genome information is being accumulated is staggering and challenging scientists’ ability to collect, process, and understand the new data. The challenge for food scientists is shifting from the goal of obtaining more genome sequence to the more difficult aim of comprehending the impact of specific gene expression patterns and then applying that information to improve the safety and nutritive value, as well as lower the food production costs.
Production of a genome sequence is only the beginning of a new road to discovery. The heart of the discovery lies in the new fields of functional and comparative genomics, along with proteomics (Fields, 2000). Agriculture, nutrition, and medicine will be impacted by these fields. The sectors that put the new information from these fields to good use will benefit immensely from new or improved products and increased safety.
Functional genomics uses various molecular tools to examine how organisms utilize their genomes (by expressing specific genes) in different situations or environments. Since most genomes have thousands of genes, this task is a little like trying to monitor all the individual conversations in a crowded football stadium at one moment in time. In essence, functional genomics documents a cell’s many “conversations.” This in turn sheds light on a web of metabolic networks, providing an unprecedented view of how a living cell carries out its many functions.
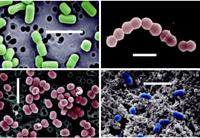
Another level of complexity is that of the protein interactions once the genes are expressed. Following the football stadium analogy, this equates to monitoring all the conversations and all the personal interactions. This is a very difficult task, but it is one of the most important issues for future food scientists. Understanding and predicting cellular responses to changing environmental stimuli will help food scientists define and, more important, exploit such responses to benefit food safety, production consistency, and flavor enhancement. For example, a deeper comprehension of the bacterial response to acid stress will aid in designing processing parameters that allow certain beneficial microorganisms, such as lactic acid bacteria (LAB) starter cultures (Fig. 1), to survive, while others, such as spoilage and pathogenic organisms, are inhibited or killed. Functional genomics can provide the tools to yield the detailed information required to meet the goals of increased consistency and safety.
So, what tools does one use to do functional genomics? Well, we start with a genome sequence, either completely or incompletely finished (known as a draft sequence). Then, we need a way to isolate the various cellular gene expression intermediates or products (analogous to the individual conversations in the football stadium) so they are each distinct and separate. This can be done in several fashions, focusing on either cellular expression of RNA or protein, respectively.
Genome arrays are the method of choice to document whole genome expression by monitoring the cellular RNA content. This is done by separately placing a snippet of DNA specific for each gene (and the corresponding RNA) within a genome onto a nylon membrane or glass slide to generate an array of signature DNA sequences (or “probes”). Cellular RNA is isolated from an organism and converted to cDNA (made from that RNA by reverse PCR) as a tag is incorporated. Often the tag is fluorescent, but other options are available. The tagged cDNA is hybridized to the genome array to produce a signal on the probe spot. Alternatively, the RNA is tagged and hybridized to the cognate DNA probe spot on the array. In this fashion, all the RNA expressed within a cell can be recorded and different samples can be compared (imagine comparing the conversations in a football stadium that occur after a touchdown to those arising after a bad call by the referee).
--- PAGE BREAK ---
Working with DNA arrays requires immense data-handling capacity. For example, an experiment with a DNA array of an entire genome, even a small one, requires approximately 2,000–2,500 individual spots with DNA probes attached to the surface. A single experiment with a single variable can produce 2,500–5,000 data points that need to be done in duplicate. Thus, a single condition will produce 5,000–10,000 discrete data points. A single experiment defines the set of mRNA gene products that were produced in a single specific condition. To make this information useful, a comparative experiment needs to be done in the same fashion, extracting RNA from a different condition. Thus, the outcome between these two experiments is 10,000–20,000 data points that need to be acquired, processed, analyzed for validity, and compared on a pairwise basis.
The individual pair-wise comparisons define the expression differences, which are then associated with a biological function to determine the metabolic difference between the test conditions. These numbers balloon by a factor of 1,000–10,000 when working with plants or animals. As one can imagine, computer science and statistics are an integral part of the analysis. How to merge these disciplines is a limiting factor at this point, but extensive work is underway.
Proteomics involves monitoring whole-cell protein expression. This approach uses a more traditional tool, two-dimensional gel electrophoresis (2D), to separate the proteins present in a cell at a specific point in time during growth. Like RNA expression, protein-expression profiles from cells immersed in different environmental conditions can be compared. The protein spots on a 2D gel that differ between conditions are eluted or cut out and digested by specific proteases, then subjected to mass spectrometry or tandem mass spectrometry. The resultant mass fingerprint is used to link the 2D gel spot to a specific gene within the genome. In this fashion, proteins expressed from the genome are catalogued and monitored and collectively known as the proteome.
Whole-cell protein expression profiling provides a fundamentally different, yet complementary, view of cellular metabolism than RNA profiling. For example, proteins may persist in a cell longer (or degrade faster) than their cognate mRNA transcripts,such that RNA analysis may not give an accurate view of the proteins available in a cell at any moment in time. Conversely, proteome analysis often misses proteins that are bound in the membrane or are not in the water phase of the cell mass. Hence, these complementary methods provide a fuller picture and understanding of cellular gene expression when used together. This process provides a wealth of information about the cell and how it responds to the environment. With these data, strategies can be formulated to modulate bacterial survival in food, as well as modify metabolic processes during storage.
Comparative genomics is a field that is emerging from the flood of available genome sequence information. As the name implies, comparative genomics examines whole genome sequences between organisms to examine the shared and disparate genes. This is essential to understand how various organisms evolved and has many applications in the realm of food science (for example, how did Escherichia coli O157:H7 evolve to fulfill the terrible role it currently plays in our food supply?). Another use of these computer analyses is the determination of elements that represent the core metabolic pathways and abilities for a genus or species.
Comparative analyses between bacterial genomes are very interesting and hold great promise to provide new practical information because their genomes are very dynamic (Hughes, 2000). Gene duplication, translocation, inversion, deletion, and horizontal transfer often mediate genome rearrangements. Presumably, such rearrangements mediate rapid strain evolution and adaptation (Guédon et al., 2000; Hughes, 2000). Comparing sequenced genomes is an excellent technique in beginning to understand genome plasticity and how it impacts the organisms associated with food processing.
Whole genome analysis is only possible if both genomes have been sequenced. However, it is also possible to use genome arrays to compare the genomes of organisms that have not been sequenced. In this case, a whole genome from organism A is fragmented into small pieces and hybridized to a genome array from the genome sequence of organism B. The amount of similarity between the two organisms is determined by the pattern of spots on the array. For example, genome arrays can be used to compare pathogenic and nonpathogenic serotypes (as is the case with E. coli K12 and E. coli O157:H7) to reveal genetic factors with a potential role in virulence.
This kind of analysis is critical to help scientists understand particular details about how genomes evolve, why organisms occupy different environmental niches, or how the strains differ in their metabolic capability. Using this approach to explore the genomes of LAB starter cultures is important, since there are too many industrially important strains to sequence them all. Thus, this type of comparative approach may be an important tool for LAB strain selection to improve fermented foods. One can also imagine that comparative genomics will provide insight into why certain pathogens are associated with specific foods.
--- PAGE BREAK ---
Recent Advances
The most dramatic advance in genomics is development of instrumentation to generate genome sequence very fast. This advancement was motivated by the need to sequence large genomes, primarily the human genome, quickly. These advances have resulted in opportunities to sequence bacterial genomes very fast—days rather than months. For example, the Joint Genome Institute (JGI), Walnut Creek, Calif., a human genome sequencing facility, sequenced the genome of Enterococcus faecium in a single day (JGI, 2000a, b). This organism is often multi-drug resistant and is commonly associated with food as a fecal contaminant. Pseudomonas fluorescens, a food–associated spoilage microbe, was also sequenced by JGI (JGI, 2000c). This sequencing capacity is an exciting development and allows many organisms with small genomes to be sequenced very quickly. Unfortunately, the ability to produce genome sequence far outpaces our ability to collect and use expression array information. However, this is no reason to stop producing genomes. Rather, it highlights the importance of selecting genomes for sequencing that can be explored with genome arrays to produce an immediate impact.
Additional advances in the miniaturization, stability, and specificity of genome arrays are being announced. In most cases, bacterial genome arrays can be done by an extension of classical membrane techniques because there are relatively few genes (~2,000 compared to ~10,000 for eukaryotes). A number of approaches with specific refinements are available to make genome arrays for use with microbial genomes. Further practical uses can be exploited when RNA/DNA extraction methods and reagents for use with food and environmental samples become available.
The mass of parallel information generated from arrays generated a need for computer programs to handle the data acquisition and analysis. Many companies are rushing to produce software to handle this need (Goodman, 2002). An extension of the need for powerful computing is the need for statistical validity and appropriate data-handling procedures.
Use of expression arrays for genome-scale experiments is interesting and in their infancy in laboratory conditions, let alone directly from food. Expression arrays offer the opportunity to determine how deletion of a single gene influences the rest of the genome expression and organism; they also play a role in defining the function of unknown genes. When expression arrays and gene knock-outs are combined with proteomics and analytical biochemistry, a robust and comprehensive picture of the cellular behavior emerges. Merging these areas can be applied to detection of spoilage and pathogenic organisms directly in the food matrix, instead of relying on cell growth on artificial media.
Microbial Genomes
The majority of bacterial genome sequencing projects have focused on clinical pathogens. Clearly, this these genome sequences are immediately useful. Worldwide, there are about 60 completed microbial genomes, according to The Institute for Genomic Research (TIGR), Rockville Md., with many more beginning at production sequence facilities such as TIGR and the Sanger Institute (TIGR, 2002; Sanger Institute, 2002). A number of foodborne pathogens are finished or are in the process of being sequenced. Additional food-associated microbe genomes are finished or being sequenced in many laboratories around the world; see the Web sites for NCBI (2002), Sanger Institute (2002), and TIGR (2002) for a complete listing of these organisms. The organisms in the public domain with direct benefit for food fermentation are Saccharomyces cerevisiae (Newes, 1997; NCBI, 2002b) and Lactococcus lactis IL1403 (Bolotin et al., 2001); however, others are on the way (see sidebar on p. 188 describing the Lactic Acid Bacteria Genome Consortium).
It quickly became obvious that the genome sequence alone is interesting, but before the sequence alone can be truly useful to agriculture and the food industry, additional information is needed about how the organism interacts with the environment at a gene expression level. This realization opened the door for the development and use of expression arrays. To date, little information is available to demonstrate the utility of this approach. Recently, Backhus et al. (2001) used the genome sequence of accharomyces cerevisiae to measure gene expression wine, highlighting the application of expression array information to improve fermented foods. No published studies are available using entire genome arrays of lactococci with this aim in mind. However, Kuipers (2001) notes the possibility that the physiology, metabolism, and genome expression regulatory mechanisms in lactococci can be now be explored in fine detail. Undoubtedly, there are groups working on this to demonstrate lactococcal functionality in food fermentations and safety.
Implications for Food Production and Processing
The future challenge for food microbiologists and food processors is to use these tools, and the resulting information, to enhance the food supply. Work in this area is just beginning and has the potential to dramatically impact how microbes are used in food processing in the future. Functional genomics has the potential to completely change our understanding of specific processing steps and how they impact organism metabolism. For example, availability of the genome sequence of a beneficial intestinal bacterium will provide a wealth of information to detect and follow that bacterium in the intestinal tract, despite the plethora of other organisms. This type of advancement will revolutionize how we define microbe gene expression, host interactions, and bacterial interaction with the environment.
--- PAGE BREAK ---
To fully appreciate the impact of these new approaches, one must think in a global and community context for cellular function. We can now define how to control (promote or hinder) gene expression, determine the genetic relatedness of organisms in a community, and examine new ecological questions with an unprecedented level of resolution. These questions have practical importance for food production and processing, especially for raw material quality and safety, successful food fermentations, and demonstrating the impact of functional foods. For example, it may now be possible to determine microbial populations on raw materials and monitor the population dynamics throughout the processing stream—all by following gene expression rather growing the cells (Cho and Tiedje, 2002). This holds promises to further refine processing steps to further inhibit bacteria during food processing.
Production of genetically modified crops and foods is common. Traditionally, a single gene has been modified to create a genetically modified organism (GMO), whether it be in a crop or an LAB starter culture. Use of genome expression arrays and functional genomics offers new tools to determine the effect of this single gene modification. It also offers a mass screening tool that can be used to determine adulteration of natural populations. In short, genomics offers a new way to assess the safety of GMOs.
Genome expression arrays will aid in defining LAB starter culture metabolism mechanisms for acid production, flavor compounds, or antimicrobial substances. This is an important use of arrays, since fermented foods account for approximately $21.5 billion for food processing (Census of Manufacturers, 1997a-e) and nearly every winery uses LAB to carry out the malolactic conversion (Fleet, 1999). For example, details defining the interaction between sugar and flavor production pathways are now possible. Additionally, regulation of acid production can be assessed with the aim of producing lactic acid faster to inhibit pathogens. Use of functional genomics in LAB starter cultures holds great promise to define and modify elusive metabolic mechanisms used by these organisms in fermented foods.
This approach to food science and food microbiology is very powerful. It will provide definitive processing techniques that promote safer foods. Agriculture and food processors are in the position to expand and exploit functional genomics for the betterment of our food supply—the ultimate aim of food scientists and food microbiologists.
Lactic acid bacteria genome consortium formed
The Lactic Acid Bacteria Genome Consortium is a collaborative group of leading scientists in lactic acid bacteria from seven universities across the United States. David Mills and Bart Weimer initiated the consortium to provide a framework of basic information and tools needed to advance the information about lactic acid bacteria. This resulted in the opportunity to obtain and apply the genome sequence of variety of bacteria associated with food fermentation for a useful outcome.
The consortium was formed with a unified focus to determine the genome sequence of a number of food-related bacteria, determine how they exert their metabolic impact via a genomics approach, understand how these bacteria evolve (i.e., mutate), and explore metabolic regulation in various environmental conditions. By understanding these questions, the consortium expects to generate an understanding of cellular metabolism and evolutionary genomics that will be used to improve food safety and enhance the quality of fermented foods.
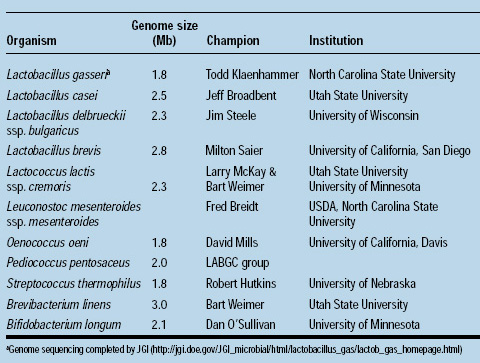 The members of the consortium and the organisms they are sequencing are listed at right. More information about the consortium is available from the authors.
The members of the consortium and the organisms they are sequencing are listed at right. More information about the consortium is available from the authors.
by Bart Weimer and David Mills
Author Weimer is Associate Professor, Dept. of Nutrition & Food Sciences, and Director, Center for Microbe Detection & Physiology, Utah State University, Logan, UT 84322-8700. Author Mills is Assistant Professor, Dept. of Enology and Viticulture, University of California, Davis, CA 95616- 8749. Send reprint requests to author Weimer.
References
Backhus, L.E., DeRisi, J., Brown, P.O., and Bisson, L.F. 2001. Functional genomic analysis of a commercial wine strain of Saccharomyces cerevisiae under differing nitrogen conditions. FEMS Yeast Res. 1: 111–125.
Bolotin, A., Wincker, P., Mauger, S., Jaillon, O., Malarme, K., Weissenbach, J., Ehrlich, S.D., and Sorokin, A. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11: 731-753.
Census of Manufacturers. 1997a. C.M.-E. 1997. Industry series: Cheese manufacturing. www.census.gov/prod/www/abs/97ecmani.html
Census of Manufacturers. 1997b. E.D.P.M.-E. 1997. Industry series: Dry, condensed, evaporated dairy product manufacturing. www.census.gov/prod/www/abs/97ecmani.html
Census of Manufacturers. 1997c. F.A.V.C.-E. 1997. Industry series: Fruit and vegetable canning. www.census.gov/prod/www/abs/97ecmani.html
Census of Manufacturers. 1997d. F.M.M.-E. 1997. Industry series: Fluid milk manufacturing. www.census.gov/prod/www/abs/97ecmani.html
Census of Manufacturers. 1997e. M.P.F.C.-E. 1997. Industry series: Meat processed from carcasses. www.census.gov/prod/www/abs/97ecmani.html.
Cho, J–C. and Tiedje, J.M. 2002. Quantitative detection of microbial genes by using DNA microarrays. Appl. Environ. Microbiol. 68: 1425–1430.
Fields, S. 2000. Proteomics in genomeland. Science 291: 1221–1224.
Fleet, G.H. 1999. Microorganisms in food ecosystems. Intl. J. Food Microbiol. 50: 101-117.
Guédon, G., Bourgoin, F., Burrus, V., Pluvinet, A., and Decaris, B. 2000. Implication of horizontal transfers in genetic polymorphism of lactic acid bacteria. Sciences des Aliments 20: 85-95.
Goodman, N. 2002. A dimsummary of microarray software. Genome Technol. 19: 58-64.
Hughes, D. 2000. Evaluating genome dynamics: The constraints on rearrangements within bacterial genomes. Genome Biology 1: reviews 0006.1–0006.8.
JGI. 2000a. JGI sequences “supergerm” genome in one day. http://jgi.doe.gov/News/news_5_9_00.htm.
JGI. 2000b. Researchers unravel genome for “superbug” bacterium using one day’s production capacity. http://jgi.doe.gov/News/news_5_11_00.html.
JGI. 2000c. Pseudomonas fluorescens genome project. http://jgi.doe.gov/JGI_microbial/html/pseudomonas/pseudo_mainpage.html.
Kuipers, O. 2001. Complete DNA sequence of Lactococcus lactis adds flavor to genomics. Genome Res. 11:673-674.
Mewes, H.W., Albermann, K., Bahr, M., Frishman, D., Gleissner, A., Hani, J., Heumann, K., Kleine, K., Maierl, A., Oliver, S.G., Pfeiffer, F., and Zollner, A. 1997. Overview of the yeast genome. Nature 387: 7-65.
NCBI. 2002a. Microbial genome sequencing projects. www.ncbi.nlm.nih.gov:80/PMGifs/Genomes/micr.html.
NCBI. 2002b. The yeast genome page. www.ncbi.nlm.nih.gov:80/cgi-bin/Entrez/map00?taxid=4932.
Sanger Institute. 2002. Microbial genome sequencing projects. www.sanger.au.uk.
TIGR. 2002. Microbial genome sequencing projects. www.tigr.org.
