Bacterial Biofilms: Formation, Prevention, and Control
FOOD SAFETY AND QUALITY
In a recent review article, Niels Høiby defined microbial biofilms as “an aggregate of microbial cells surrounded by a self-produced polymer matrix” (Høiby 2017). Discussing their history, he said, “The observation of biofilms is old,” “since both Leeuwenhoek and Pasteur have described the phenomenon.” In the late 1600s, Anthony van Leeuwenhoek (1632–1723) wrote to the Royal Society of London about his observations of the plaque between his teeth, which he made with his primitive microscopes. In the late 1800s, Louis Pasteur (1844–1895) observed aggregates of bacterial cells as the source of wine becoming vinegar. ZoBell and Allen (1935) introduced the term biofilm in environmental microbiology in discussing the significance of marine bacteria in the fouling of submerged surfaces. In the early 1970s, Høiby himself observed aggregates of Pseudomonas aeruginosa cells in chronically infected cystic fibrosis patients, by microscopic examination of mucus and lung tissue, initiating the concept of “biofilm infection.” The term biofilm was introduced into medical microbiology and medicine by Nickel et al. (1985), who demonstrated increased antimicrobial resistance of bacterial cells growing in biofilms as compared with their planktonic counterparts.
Relevance to the Food Industry
In food-related environments, including the meat, dairy, and fresh produce sectors, surfaces and equipment are habitually populated by microorganisms forming biofilms (Pometto and Demirci 2015). Biofilms can slough off, move to surrounding environments, elevate intermediary and final product microbial loads, and reduce shelf life. Biofilms can also serve as a source of cross-contamination, consequently diminishing effectiveness of food processing technologies, compromising microbial food quality and food safety, and leading to sizable economic losses. Microbial cells in biofilms are quite well protected from adverse environmental conditions, including sanitizing agents. Thus, prevention and control of biofilms by the food industry is critical and challenging.
Biofilms are formed by spoilage microorganisms (e.g., Pseudomonas spp.) and pathogens, including Aeromonas hydrophila, Bacillus cereus, Campylobacter jejuni, Cronobacter sakazakii, Escherichia coli O157:H7, Listeria monocytogenes, Salmonella spp., Shigella spp., Staphylococcus aureus, Vibrio cholera, and Yersinia spp. (Bhunia 2018, Matthews et al. 2017, Pometto and Demirci 2015). Also formed by beneficial microorganisms, biofilms can be advantageous. They may be used to improve human health via the the gut microbiota, increase the quality and the yield of food fermentations, produce value-added products through fermentation, develop biotechnological applications for improving food safety and quality, and manage industrial waste.
Formation
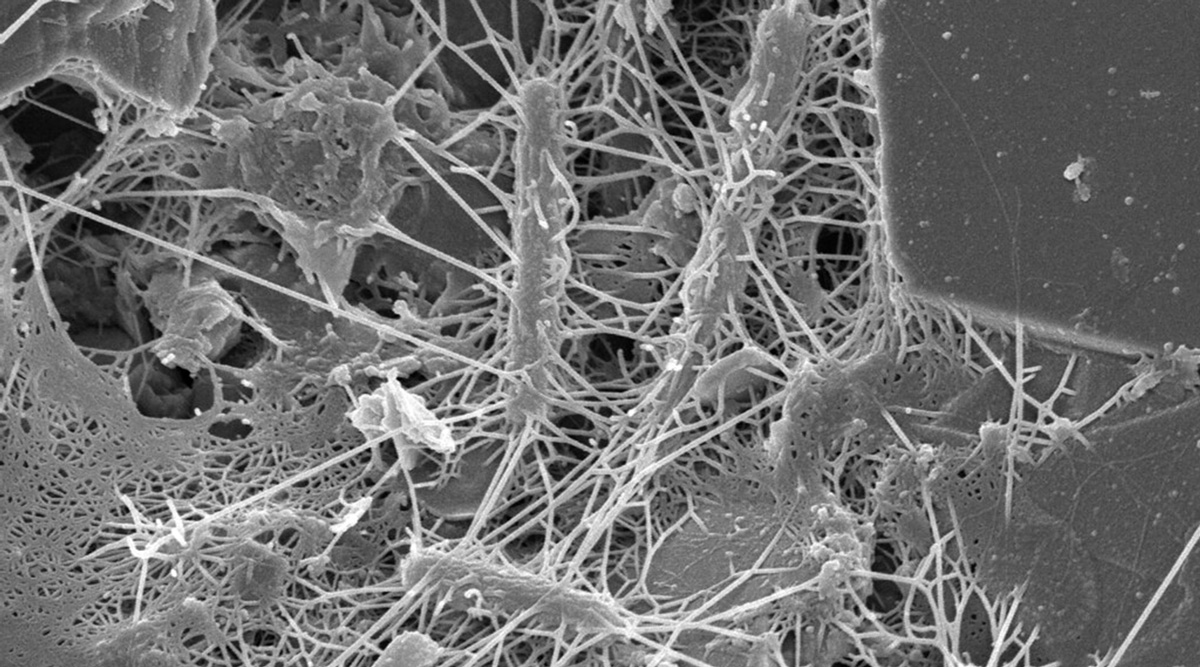
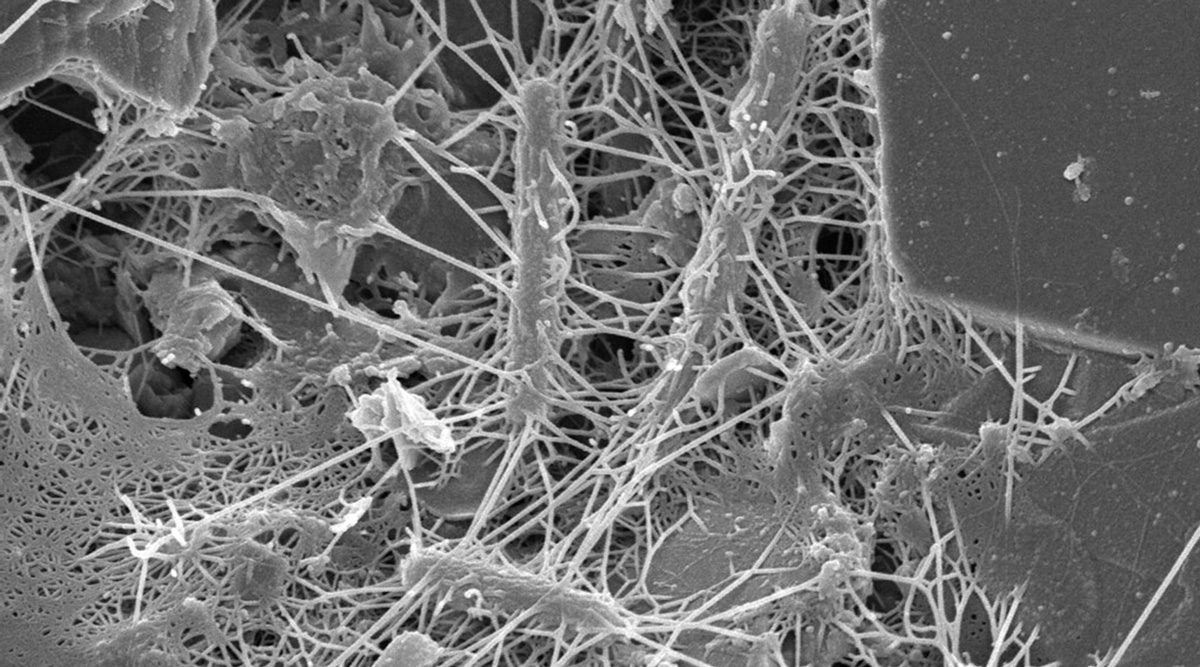
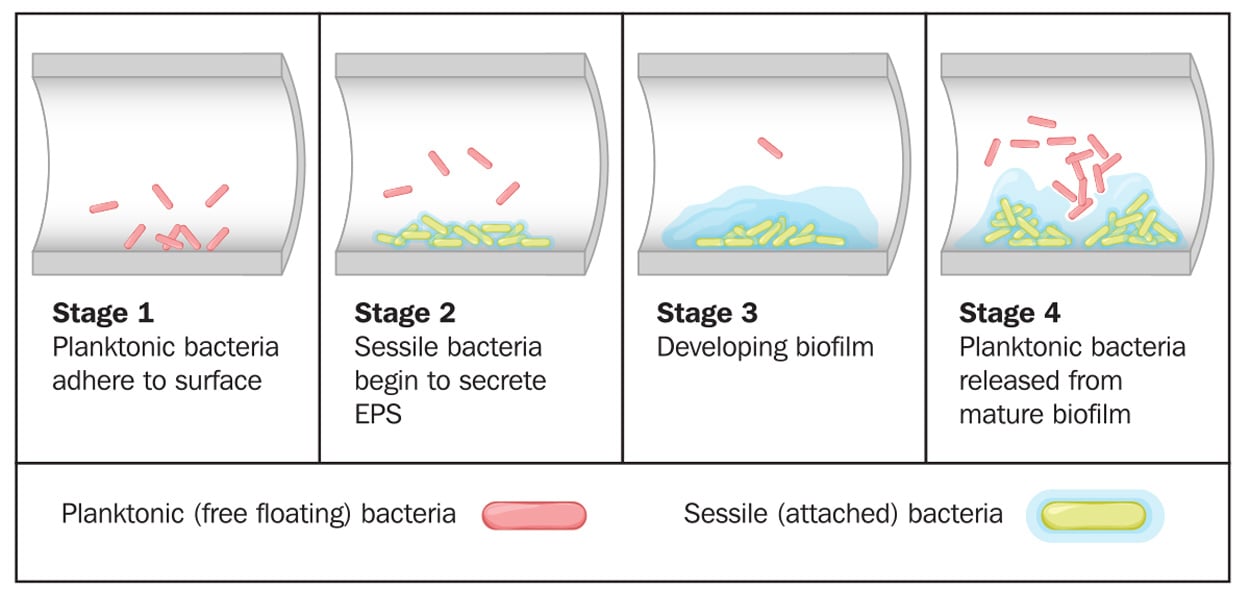
Biofilms can form on any type of surface (e.g., plastic, metal, glass, wood, or food) that is preconditioned by food particles or substrates. Biofilms may be formed by a single microorganism or a group of microorganisms, through a complex multistage process. Described by Winkelströter et al. (2014) and McLandsborough (2015), the stages of formation are as follows: initial transfer/adhesion/attachment of planktonic microbial cells to the surface, production of extracellular polymeric substance (EPS, also known as a glycocalyx), microcolony formation, and maturation into a 3D-biofilm. These stages may be followed by detachment/dispersion/recolonization of microbial cells involving new preconditioned surfaces. The first stage in attachment to surfaces is reversible sorption, thought to be related to van der Waals forces (or other physicochemical factors). The second stage deals with irreversible attachment via EPS production, which anchors the cells and protects them against environmental stresses. Many factors can influence microbial attachment, including characteristics of a surface, phase and temperature of microbial growth, and microbial motility. This stage is followed by the formation of microcolonies with well-defined boundaries, allowing fluid channels to run through the biofilm. Quorum sensing, known as microbial cell-to-cell communication, plays a role in the formation of microcolonies and organization of fluid channels.
Prevention and Control
The structural and physiological features of microbial cells within biofilms, such as reduced diffusion, anaerobic growth, reduced growth rate, and the viable but nonculturable (VBNC) state, cause them to be significantly more resistant to adverse environmental conditions than their planktonic counterparts. Use of various sanitizers and biocides in cleaning and sanitation is not sufficient to remove cells within biofilms. Proper cleaning is important for their removal and eradication before they become recalcitrant in mature biofilms (Matthews et al. 2017).
There has been a great deal of interest to develop innovative, efficient, and inexpensive biofilm inhibition and removal strategies. One promising approach uses bio-nanotechnology to modify surface materials. These include developments inspired by naturally occurring antimicrobial surfaces and substances (e.g., antimicrobial peptides), generation of artificial antimicrobial surfaces with both traditional and advanced surface modification techniques (e.g., via use of silver, gold, or titanium dioxide, and physical topographical modifications involving functionalization, derivatization, and polymerization) (Birkenhauer and Neethirajan 2015).
Nonthermal atmospheric plasmas are an innovative control technology of interest to the food industry due to their contact-free, waterless nature and their effectiveness against a wide range of microorganisms (Niemira et al. 2014, Puligundla and Mok 2017, Ziuzina et al. 2015a, b). Other inactivation technologies being investigated and applied are photodynamic inactivation using pulsed ultraviolet light (Montgomery and Banarjee 2015), steam heating (Ban and Kang 2016), light (405 nm) (McKenzie et al. 2013), ozone (Nicholas et al. 2013), ultrasound (Axelsson et al. 2013), and gaseous chlorine dioxide (Nam et al. 2014).
Development of new environmentally friendly (“green”) sanitizers capable of removing biofilms in food processing environments is growing in research interest. Enzymes such as alpha-amylase, beta-glucanase, DNase, proteinase K, and lipases are considered in formulations of enzymatic detergents due to their ability to degrade the EPS (Araújo et al. 2017, Brown et al. 2015, Nguyen and Burrows 2014, Seghal Kiran et al. 2014, Zetzmann et al. 2015). Plant-based compounds, such as essential oils, are also being considered in new “green” sanitizer formulations (Ashraf et al. 2014).
Use of microorganisms such as bacteriocin-producing lactic acid bacteria and their metabolites for competitive exclusion or inactivation of undesirable microorganisms is an area of research. A variety of bacteriocins, including enterocin AS-48 (Caballero Gómez et al. 2013), enterocin B3A-B3B (Al-Seraih et al. 2017), lichenicidin (Mathur et al. 2018), nisin and nisin derivatives (Field et al. 2016), sonorensin (Chopra et al. 2015), and subtilomycin (Mathur et al. 2018) are effective. Other bacterial metabolites, including the biosurfactant lichenysin (Coronel-León et al. 2016), endoglycosidases such as glycosyl hydrolase (Yu et al. 2015), and unsaturated fatty acids such as cis-2-decenoic acid (Sepehr et al. 2014) are also useful for biofilm prevention and control.
Lytic bacteriophages have been used as biocontrol agents against biofilms of specific microorganisms, such as C. sakazakii (Enderson et al. 2017), E. coli O157:H7 (Sadekuzzaman et al. 2017), L. monocytogenes (Nannapaneni and Soni 2015), P. aeruginosa (Shafique et al. 2017), Salmonella spp. (Islam et al. 2019, Sadekuzzaman et al. 2018), and S. aureus (González et al. 2017). Moye et al. (2018) comprehensively reviewed bacteriophage applications for food production and processing. They indicated that in recent years, several bacteriophages against a number of specific foodborne pathogenic bacteria are considered “approved” by the U.S. Food and Drug Administration and U.S. Department of Agriculture.
Need for Additional Research
Biofilms affect disparate processes in our lives, from oral hygiene, to safe water and food, bioremediation of toxic compounds, and wastewater treatment. Although biofilms have received a great deal of attention over the years, there continues to be a need for additional research. In the context of the food system, better understanding of biofilm structure, microbial physiology within biofilms, and microbial interactions (e.g., quorum sensing) would make positive contributions to effective biofilm prevention and control. A number of conventional and modern techniques have been used to detect and evaluate microorganisms in biofilms in vitro. Development of innovative techniques for assessing complex biofilm communities in food processing environments would allow in situ characterization of biofilms, and lead to successful prevention and control. A plethora of physical, chemical, and biological approaches capable of preventing biofilm formation and/or removing them exists. Nevertheless, research into simultaneous use of two or more of these approaches in combination would enhance efficacy.
REFERENCES
Al-Seraih, A., Y. Belguesmia, J. Baah, et al. 2017. “Enterocin B3A-B3B Produced by LAB Collected from Infant Feces: Potential Utilization in the Food Industry for Listeria monocytogenes Biofilm Management.” Antonie van Leeuwenhoek 110: 205–19. doi:10.1007/s10482-016-0791-5.
Araújo, P. A., I. Machado, A. Meireles, et al. 2017. “Combination of Selected Enzymes With Cetyltrimethylammonium Bromide in Biofilm Inactivation, Removal and Regrowth. Food Res. Int. 95: 101–107. doi:10.1016/j.foodres.2017.02.016.
Ashraf, M. A., S. Ullah, I. Ahmad, et al. 2014. “Green Biocides, a Promising Technology: Current and Future Applications to Industry and Industrial Processes.” J. Sci. Food Agric. 94(3): 388–403. doi:10.1002/jsfa.6371.
Axelsson, L., A. Holck, I. Rud, et al. 2013. “Cleaning of Conveyor Belt Materials Using Ultrasound in a Thin Layer of Water.” J. Food Prot. 76(8): 1401–7. doi:10.4315/0362-028X.JFP-12-563.
Ban, G.-H. and D.-H. Kang. 2016. “Effect of Sanitizer Combined with Steam Heating on the Inactivation of Foodborne Pathogens in a Biofilm on Stainless Steel.” Food Microbiol. 55: 47–54. doi:10.1016/j.fm.2015.11.003.
Bhunia, A. 2018. Foodborne Microbial Pathogens Mechanisms and Pathogenesis. 2nd ed. New York: Springer.
Birkenhauer, E. and S. Neethirajan. 2015. “Prevention and Control of Biofilms in the Food Industry and Bio-Nanotechnology Approaches.” Chpt. 4 in Biofilms in the Food Environment, edited by A. L. Palmetto III and A. Demirci. IFT Press. Ames, Iowa: Wiley Blackwell.
Brown, H. L., K. Hanman, M. Reuter, et al. 2015. “Campylobacter jejuni Biofilms Contain Extracellular DNA and are Sensitive to DNase I Treatment. Front. Microbiol. 6: 699. doi:10.3389/fmicb.2015.00699.
Caballero Gómez, N., H. Abriouel, M. J. Grande, et al. 2013. “Combined Treatments of Enterocin AS-48 with Biocides to Improve the Inactivation of Methicillin-Sensitive and Methicillin-Resistant Staphylococcus aureus Planktonic and Sessile cells.” Int. J. Food Microbiol. 163(2–3): 96–100. doi:10.1016/j.ijfoodmicro.2013.02.018.
Chopra, L., G. Singh, K. K. Jena, et al. 2015. “Sonorensin: A New Bacteriocin with Potential of an Anti-Biofilm Agent and a Food Biopreservative. Sci. Rep. 5(1): 13412. doi:10.1038/srep13412.
Coronel-León, J., A. M. Marqués, J. Bastida, et al. 2016. “Optimizing the Production of the Biosurfactant Lichenysin and its Application in Biofilm Control.” J. Appl. Microbiol. 120(1): 99–111. doi:10.1111/jam.12992.
Endersen, L., C. Buttimer, E. Nevin, et al. 2017. “Investigating the Biocontrol and Anti-Biofilm Potential of a Three Phage Cocktail Against Cronobacter sakazakii in Different Brands of Infant Formula.” Int. J. Food Microbiol. 253: 1–11. doi:10.1016/j.ijfoodmicro.2017.04.009.
Field, D., R. O’Connor, P. D. Cotter, et al. 2016. “In Vitro Activities of Nisin and Nisin Derivatives Alone and in Combination with Antibiotics Against Staphylococcus Biofilms.” Front. Microbiol. 7: 508. doi:10.3389/fmicb.2016.00508.
González, S., L. Fernández, A. B. Campelo, et al. 2017. “The Behavior of Staphylococcus aureus Dual-Species Biofilms Treated with Bacteriophage phiIPLA-RODI Depends on the Accompanying Microorganism.” Appl. Environ. Microbiol. 83(3): e02821–16. doi:10.1128/AEM.02821-16.
Høiby, N. 2017. “A Short History of Microbial Biofilms and Biofilm Infections.” APMIS 125(4): 272–75. doi:10.1111/apm.12686.
Islam, M. S., Y. Zhou, L. Liang, et al. 2019. “Application of a Phage Cocktail for Control of Salmonella in Foods and Reducing Biofilms.” Viruses 11(9): 841. doi:10.3390/v11090841.
Mathur, H., D. Field, M. C. Rea, et al. 2018. “Fighting Biofilms with Lantibiotics and Other Groups of Bacteriocins.” npj Biofilms Microbiomes 4(1): 9. doi:10.1038/s41522-018-0053-6.
Matthews, K. R., K. E. Kniel, T. J. Montville. 2017. Food Microbiology: An Introduction. Washington, DC: ASM Press.
McKenzie, K., M. Maclean, I. V. Timoshkin, et al. 2013. “Photoinactivation of Bacteria Attached to Glass and Acrylic Surfaces by 405 nm Light: Potential Application for Biofilm Decontamination.” Photochem. Photobiol. 89(4): 927–35. doi:10.1111/php.12077.
McLandsborough, L. 2015. “Current Knowledge and Perspectives on Biofilm Formation and Remediation.” Chpt. 1 in Biofilms in the Food Environment, edited by A. L. Palmetto III and A. Demirci. IFT Press. Ames, Iowa: Wiley Blackwell.
Montgomery, N. L. and P. Banerjee. 2015. “Inactivation of Escherichia coli O157:H7 and Listeria monocytogenes in Biofilms by Pulsed Ultraviolet Light.” BMC Res. Notes 8(1): 235. doi:10.1186/s13104-015-1206-9.
Moye, Z. D., J. Woolston, A. Sulakvelidze. 2018. “Bacteriophage Applications for Food Production and Processing.” Viruses 10(4): 205. doi:10.3390/v10040205.
Nam, H., H.-S. Seo, J. Bang, et al. 2014. “Efficacy of Gaseous Chlorine Dioxide in Inactivating Bacillus cereus Spores Attached to and in a Biofilm on Stainless Steel.” Int. J. Food Microbiol. 188: 122–27. doi:10.1186/s13104-015-1206-9.
Nannapaneni, R. and K. A. Soni. 2015. “Use of Bacteriophages to Remove Biofilms of Listeria monocytogenes and Other Foodborne Bacterial Pathogens in the Food Environment.” Chpt. 5 in Biofilms in the Food Environment, edited by A. L. Palmetto III and A. Demirci. IFT Press. Ames, Iowa: Wiley Blackwell.
Nguyen, U. T. and L. L. Burrows. 2014. “DNase I and Proteinase K Impair Listeria monocytogenes Biofilm Formation and Induce Dispersal of Pre-Existing Biofilms.” Int. J. Food Microbiol. 187: 26–32. doi:10.1016/j.ijfoodmicro.2014.06.025.
Nicholas, R., P. Dunton, A. Tatham, et al. 2013. “The Effect of Ozone and Open Air Factor on Surface-Attached and Biofilm Environmental Listeria monocytogenes.” J. Appl. Microbiol. 115(2): 555–64. doi:10.1111/jam.12239.
Nickel, J. C., I. Ruseska, J. B. Wright, et al. 1985. “Tobramycin Resistance of Cells of Pseudomonas aeruginosa 274 Growing as a Biofilm on Urinary Catheter Material.” APMIS 27: 619-24.
Niemira B. A., G. Boyd, J. Sites. 2014. “Cold Plasma Rapid Decontamination of Food Contact Surfaces Contaminated with Salmonella Biofilms.” J. Food Sci. 79(5): M917–22. doi:10.1111/1750-3841.12379.
Pometto III, A. L. and A. Demirci. 2015. Biofilms in the Food Environment. IFT Press. Ames, Iowa: Wiley Blackwell.
Puligundla, P. and C. Mok. 2017. Potential Applications of Nonthermal Plasmas Against Biofilm-Associated Micro-Organisms in Vitro.
J. Appl. Microbiol. 122(5): 1134–48. doi:10.1111/jam.13404.
Sadekuzzaman, M., S. Yang, M. F. R. Mizan, et al. 2017. “Reduction of Escherichia coli O157:H7 in Biofilms Using Bacteriophage BPECO 19. J. Food Sci. 82(6): 1433–42. doi:10.1111/1750-3841.13729.
Sadekuzzaman, M., M. F. R. Mizan, S. Yang, et al. 2018. “Application of Bacteriophages for the Inactivation of Salmonella spp. in Biofilms.” Food Sci. Technol. Int. 24(5): 424–33. doi: 10.1111/1750-3841.13729.
Seghal Kiran, G., A. Nishanth Lipton, J. Kennedy, et al. 2014. “A Halotolerant Thermostable Lipase From the Marine Bacterium Oceanobacillus sp. PUMB02 With an Ability to Disrupt Bacterial Biofilms. Bioengineered 5(5): 305–18. doi:10.4161/bioe.29898.
Sepehr, S., A. Rahmani-Badi, H. Babaie-Naiej, et al. 2014. “Unsaturated Fatty Acid, cis-2-Decenoic Acid, in Combination with Disinfectants or Antibiotics Removes Pre-Established Biofilms Formed by Food-Related Bacteria.” PLOS ONE 9(7): e101677. doi:10.1371/journal.pone.0101677.
Shafique, M., I. A. Alvi, Z. Abbas, et al. 2017. “Assessment of Biofilm Removal Capacity of a Broad Host Range Bacteriophage JHP Against Pseudomonas aeruginosa.” APMIS 125(6): 579–84. doi:10.1111/apm.12691.
Winkelströter L. K., F. B. Teixeira, E. P. Silva, et al. 2014. “Unraveling Microbial Biofilms of Importance for Food Microbiology.” Microb. Ecol. 68(1): 35–46. doi:10.1007/s00248-013-0347-4.
Yu, S., T. Su, H. Wu, et al. 2015. “PslG, a Self-Produced Glycosyl Hydrolase, Triggers Biofilm Disassembly by Disrupting Exopolysaccharide Matrix.” Cell Res. 25(12): 1352–67.
Zetzmann, M., M. Okshevsky, J. Endres, et al. 2015. “DNase-Sensitive and -Resistant Modes of Biofilm Formation by Listeria monocytogenes.” Front. Microbiol. 6: 1428. doi:10.3389/fmicb.2015.01428.
Ziuzina, D., D. Boehm, S. Patil, et al. 2015a. “Cold Plasma Inactivation of Bacterial Biofilms and Reduction of Quorum Sensing Regulated Virulence Factors.” PLOS ONE 10(9): e0138209. doi:10.1371/journal.pone.0138209.
Ziuzina, D., L. Han, P. J. Cullen, et al. 2015b. “Cold Plasma Inactivation of Internalised Bacteria and Biofilms for Salmonella enterica serovar Typhimurium, Listeria monocytogenes and Escherichia coli.” Int. J. Food Microbiol. 210: 53–61. doi:10.1016/j.ijfoodmicro.2015.05.019.
ZoBell, C. E. and E. Allen. 1935. “The Significance of Marine Bacteria in the Fouling of Submerged Surfaces.” J. Bacteriol. 29: 239–51.