Determining Infectious Doses of Foodborne Illness Agents
FOOD SAFETY AND QUALITY
The incidence of foodborne disease continues to increase in the United States, with infections from five of eight tracked pathogens rising, according to a report from the Centers for Disease Control and Prevention (CDC) comparing data from 2016–2018 with 2019 numbers (Tack et al. 2020). CDC data show that approximately one in six Americans (or 48 million people) get sick from foodborne disease each year, 128,000 of them are hospitalized, and 3,000 die from these illnesses.
Foodborne illnesses are classified into three types: intoxication, toxicoinfection, and infection. Intoxications occur when preformed toxins are ingested and result in rapid onset of symptoms. Toxicoinfections are foodborne illnesses caused by the toxins produced inside the host by live pathogens after food ingestion. Foodborne infections take place when live and generally invasive pathogenic microorganisms are ingested and cause severe tissue damage in the host.
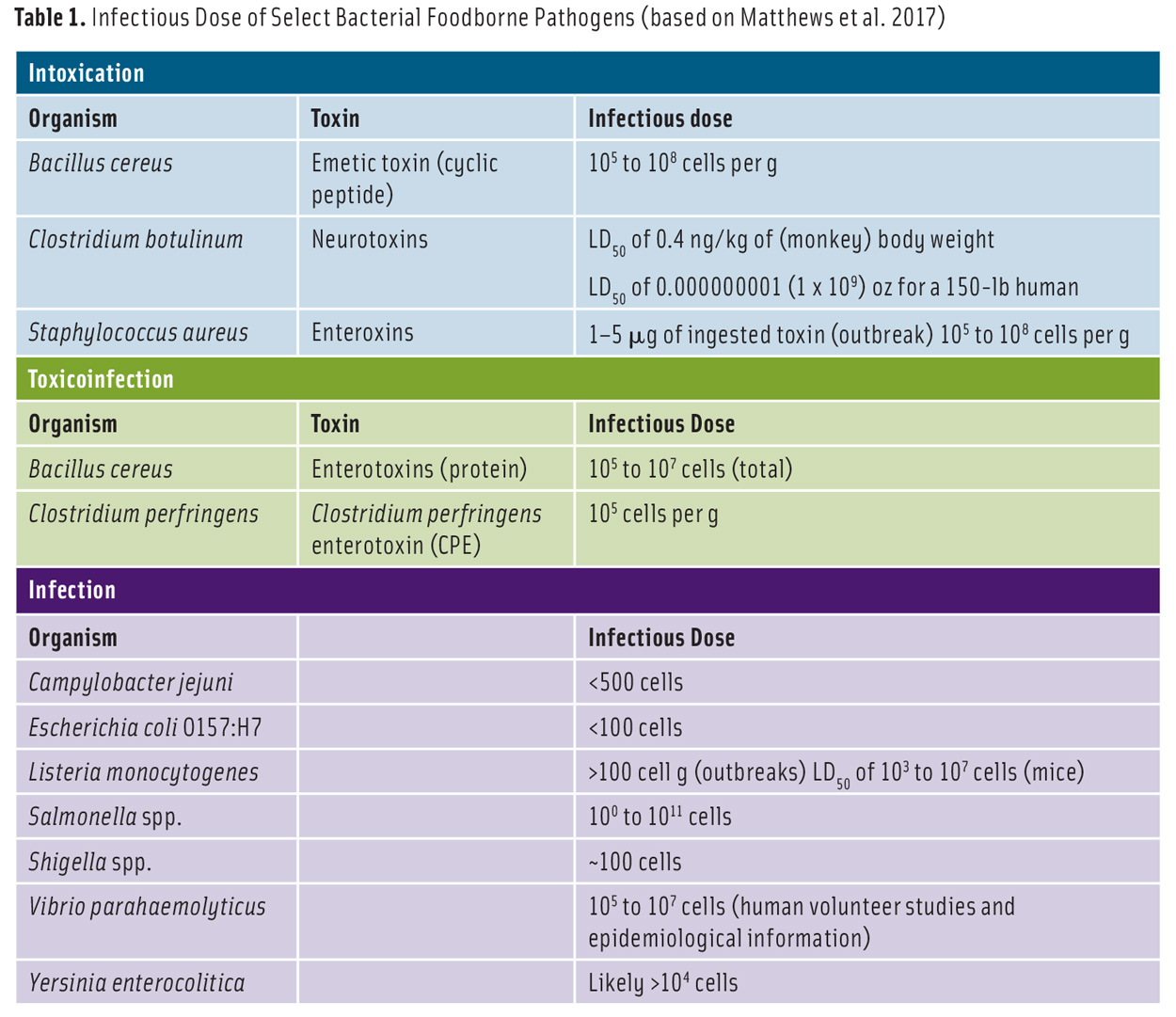
Infectivity can be defined as the likelihood that an agent will infect a host upon exposure. The infectious dose of a foodborne pathogen or toxin (Table 1) varies depending on the immunological health status of the host and the infectivity of the microorganism. People with high gastric pH or who are undergoing antibiotic therapy tend to be more susceptible to foodborne infections because antibiotics lessen the natural microbiota in the intestine. If the pathogen is consumed with fluid or viscous food, which navigates the stomach rapidly, or food that quickly neutralizes the stomach acid, the infectious dose decreases.
Infectious Dose Response Assessment
Infectious dose response assessment can be based on foodborne illness outbreaks, human clinical experiments, in vitro model of the human intestinal tract, and animal data. In general, the number of cases in outbreaks is usually well known, but the number of pathogens in the food (i.e., the dose) is not always known. So data from outbreaks lead to meaningful dose response assessment for a limited number of pathogens.
In addition, very low numbers of select foodborne pathogens can pose a substantial threat to subsets of immunocompromised individuals. This strengthens the concept that the infectious dose for such organisms should be considered as a function of the population exposed instead of a single infectious dose for all. Finally, the long time between the exposure to a select foodborne pathogen and the onset of foodborne illness can make it extremely challenging to determine accurate dose information from outbreak data.
Although humans are the ultimate model to study foodborne illness agents, biosafety and bioethics concerns preclude this. However, human volunteers have been used in some studies, such as human challenge studies conducted with two strains of Campylobacter jejuni in the classical study by Black et al. (1988).
Gastrointestinal Tract Model Systems
Foodborne pathogenic bacteria should be able to pass various barriers—esophagus (saliva), stomach (low pH and pepsin), and small intestine (bile, enzymes, and microflora)—in the human host prior to reaching a suitable site for colonization. This means that in vitro models that simulate the human gastrointestinal tract can be used to study infectious dose assessment in humans (Wijnands et al. 2017).
Animal Models
Many factors play roles in selecting an animal model for host-foodborne illness agent interactions: Pathogens must infect animals through the same route as they do for humans, display a similar colonization pattern, and show the same/similar virulence as they do in humans. Due in part to the specificity of human host-foodborne pathogen relationships, translation of animal model results to the human host can be challenging.
Rodents—gerbils, guinea pigs, ferrets, hamsters, mice, rabbits, and rats—are the most frequently used animal models. An oral or intragastric route usually is recommended to assess pathogenicity of a foodborne illness agent. The virulence measurement is mostly conveyed as the infectious dose (ID) or the lethal dose (LD). The infectious dose 50 (ID50) is defined by the number (or concentration) of a pathogen required to infect 50% of the animals, while the lethal dose 50 (LD50) is defined by the number (or concentration) required to kill 50% of the animals.
Occasionally animals are unresponsive to human pathogens. Under these circumstances, immunocompromised or immunodeficient animals can be used (Jiminez et al. 2015). While subjecting animals to gamma irradiation leads to immunocompromised animals, the immunodeficient animals are genetically engineered (i.e., transgenic). Both categories of animals are expensive to purchase and handle because they require a pathogen-free environment. Suckling mouse model, for example, has been used to study the pathogenesis of diarrheagenic microorganisms such as Vibrio cholerae and enterotoxigenic E. coli (Giannella 1976). In this model, microorganisms (or the toxin preparations) of interest are orally administered in suckling mice (2–4 days old), and the mice are then sacrificed and examined for fluid accumulation in the gut after 4–24 hr. The enterotoxic effect of the toxins is determined based on the ratio of gut weight to the body weight.
Animal Organs
Ligated intestinal loop (LIL) (Caserta et al. 2011) and embryonated eggs (Andersson et al. 2015) are the organs usually involved in foodborne illness agent investigations. Intestinal inflammation and fluid secretion by Bacillus, Clostridium, Escherichia, Salmonella, Shigella, and Vibrio species (and their toxins) have been studied using the LIL assay, with the LIL from calves, chickens, guinea pigs, mice, piglets, rabbits, and rats.
In the assay, animals who are deprived of food and water overnight are anesthetized. Then, a slight incision is created in the abdomen, a portion of the small intestine is pulled out from the abdominal cavity, and knots are placed in 2–3 cm intervals, using strings, to create loops. Experimental and control samples are injected into the loops, the intestine is put back in the abdomen, the skin incision is closed, and after 18–24 hr, the intestine is examined for fluid accumulation (by the diarrheagenic toxin). In the case of the embryonated chicken egg, 12- to 14-day-old embryonated hen’s eggs are injected aseptically with microorganisms of interest—Listeria monocytogenes, Yersinia enterocolitica, etc.—and incubated for 3–4 days. The infectiveness of the test organisms is shown by the death of embryos.
Zebrafish, Insects, and Nematodes
As substitutes for laboratory animal models, zebrafish (Danio rerio, Shan et al. 2015), fruit fly (Drosophila melanogaster, Chambers et al. 2012), larvae of the greater wax moth (Galleria mellonella, Mukherjee et al. 2010), and nematodes (Caenorhabditis elegans, Thomsen et al. 2006) are gaining recognition.
Zebrafish embryo, used in the first seven days after the eggs are deposited, is a model organism to study the function of the vertebrate immune system during microbial infection. The microorganism of interest is injected to the zebrafish embryo, which is transparent, allowing imaging and manipulation using a microscope. This model has been used to study foodborne pathogens such as Salmonella enterica serovar Typhimurium. To maintain zebrafish, specific facilities as well as the know-how for microinjection are required.
The greater wax worm, Galleria mellonella, has been described as a suitable model for the virulence assessment of various pathogens, including Listeria monocytogenes. Larvae injected with Listeria monocytogenes are usually incubated and monitored over several days for the time needed to kill 50% of larvae (LT50) and to determine change of bacterial population in Galleria mellonella several hours post-incubation. The advantages associated with the model include affordability, availability, and lack of ethical issues and regulatory restrictions. Among the major disadvantages of the model are the narrow range (106 CFU/larvae) for doses that allow infectious dose assessments and the exceptionally low virulence observed with select outbreak strains.
REFERENCES
Andersson, C., J. Gripenland, J. Johansson. 2015. “Using the Chicken Embryo to Assess Virulence of Listeria monocytogenes and to Model Other Microbial Infections.” Nat. Protoc. 10(8): 1155–1164. doi: 10.1038/nprot.2015.073.
Black, R. E., M. M. Levine, M. L. Clements, et al. 1988. “Experimental Campylobacter jejuni Infection in Humans.” J. Infect. Dis. 157(3): 472–479. https://doi.org/10.1093/infdis/157.3.472.
Caserta, J. A., S. L. Robertson, J. Saputo, et al. 2011. “Development and Application of a Mouse Intestinal Loop Model to Study the in vivo Action of Clostridium perfringens Enterotoxin.” Infect. Immun. 79(8): 3020-3027. doi: 10.1128/IAI.01342-10.
Chambers, M. C., K. H. Song, D. S. Schneider. 2012. “Listeria monocytogenes Infection Causes Metabolic Shifts in Drosophila melanogaster.” PLoS ONE 7(12): e50679. https://doi.org/10.1371/journal.pone.0050679.
Giannella, R. A. 1976. “Suckling mouse model for detection of heat-stable Escherichia coli enterotoxin: characteristics of the model.” Infect. Immun. 14(1): 95-9. doi: 10.1128/IAI.14.1.95-99.1976.
Jiminez, J. A., T. C. Uwiera, G. Douglas Inglis, et al. 2015. “Animal Models to Study Acute and Chronic Intestinal Inflammation in Mammals.” Gut Pathog. 7(29). https://doi.org/10.1186/s13099-015-0076-y.
Matthews, K. R., K. E. Kniel, T. J. Montville. 2017. Food Microbiology: An Introduction. Washington, DC: ASM Press.
Mukherjee, K., B. Altincicek, T. Hain, et al. 2010. “Galleria mellonella as a Model System for Studying Listeria Pathogenesis.” Appl. Environ. Microbiol. 76(1): 310–317. doi: 10.1128/AEM.01301-09.
Shan, Y., C. Fang, C. Cheng, et al. 2015. “Immersion Infection of Germ-free Zebrafish with Listeria monocytogenes Induces Transient Expression of Innate Immune Response Genes.” Front. Cell. Infect. Microbiol. 6: 373. doi: 10.3389/fmicb.2015.00373.
Tack, D. M., L. Ray, P. M. Griffin, et al. 2020. “Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2016–2019.” Morb. Mortal. Wkly. Rep. 69: 509–514. doi: http://dx.doi.org/10.15585/mmwr.mm6917a1external icon.
Thomsen, L. E., S. S. Slutz, M.-W. Tan, et al. 2006. “Caenorhabditis elegans Is a Model Host for Listeria monocytogenes.” J. Appl. Environ. Microbiol. 72(2): 1700-1701. doi: 10.1128/AEM.72.2.1700-1701.2006.
Wijnands, L. M., P. F. M. Teunis, A. F. A. Kuijpers, et al. 2017. “Quantification of Salmonella Survival and Infection in an In vitro Model of the Human Intestinal Tract as Proxy for Foodborne Pathogens.” Front. Cell. Infect. Microbiol. 8: 1139. doi: 10.3389/fmicb.2017.01139.