Dormant Microbes: Research Needs
IFT’s inaugural Research Summit assessed the state of the science and critical research needs regarding spores and other dormant microorganisms.
The Institute of Food Technologists convened its first Research Summit on January 12–14 in Orlando, Fla., to facilitate an in-depth interchange of information among world-renowned investigators and other scientists conducting basic and applied research on the physiology of bacterial spores and other dormant microbes, rapid quantitative measurement methods, and advanced techniques for evaluating microbial viability. The main goal of the interchange was to ultimately identify the needs for further research in these areas.
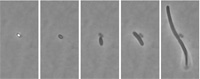
During the summit, “Rapid Measurement of Bacterial Spores and Other Dormant, Difficult-to-Measure Microorganisms—Emphasizing Quantitative Methods and Nanotechnology,” 35 scientists from six countries and multiple areas of research interests participated in dynamic exchanges and reached consensus on specific research needs.
State of the Science
Two keynote addresses, one by Grahame Gould, formerly of Unilever Research, Bedford, UK, and the other by Peter Setlow of the University of Connecticut Health Center, delved into what is known and what remains elusive in our understanding of spore physiology and the underlying system of dormancy and resistance, sporulation, initiation, germination, outgrowth, and mechanisms of inactivation.
“The spore itself has a preservation system that is par excellence, with a shelf life of thousands to millions of years,” Gould said. “If we understood more about the way a spore is put together, then maybe we would better understand how to deal with it. Maybe we could even copy some of the mechanisms employed by the spore to better preserve ambient stable foods.” More specifically, he said, the barrier to further progress is our inadequate understanding of the basic mechanisms of spore resistance, dormancy, germination, extreme dormancy (viable but nonculturable state, VBNC), and heterogeneity. Setlow said this same barrier existed 30 years ago. While considerable progress has been made in understanding the modes of action for some bacterial inactivation methods, such as irradiation, we still do not know how heat kills spores, “which is disgraceful,” Gould opined.
Key areas needing investigation, Gould said, are: (1) How does the cortex peptidoglycan act at the architectural/molecular/material science level? (2) What is the physical status of the protoplast; e.g., is it in glassy state? (3) How does calcium dipicolinate (CaDPA) contribute to dormancy and resistance? (4) How do alkyl amines and CaDPA act in germination? (5) What are the exploitable new hurdle techniques (e.g., nisin in new combinations)? (6) What are the most profitable approaches for food science/technology/industry/consumers? For most of these topics, well-integrated, multidisciplinary research collaborations offer the best chance of success.
Regarding spore killing, Setlow said it is essential to study the initial event; and when investigating the kinetics of killing in particular, it is crucial to evaluate the inactivation of 90– 95% of the total population so that killing events can be related to the specific chemical changes in the spore. Setlow also expressed the concern that studies on spore killing, particularly with Bacillus subtilis, often use mutants. While mutants can be useful, they can be misleading, since there is evidence that some mutants exhibit altered spore resistance because of alterations in global gene expression caused by the mutation and not because of direct, specific effects on spore resistance.
Other principles that Setlow highlighted and concerns he expressed are: (1) Because sporulation conditions can alter spore resistance, might this alter the mechanism of spore killing? (2) Most work on killing mechanisms has been done with B. subtilis; do all species, in particular ones of applied interest, behave similarly? (3) Can multiple killing mechanisms contribute to inactivation of a spore population? (4) Are spores dead or just “superdormant?”
Several additional presentations followed the keynote addresses before participants broke into smaller groups to reflect on what is known vs unknown about spores and the implications for future research. Highlights of the presentations are briefly captured below.
--- PAGE BREAK ---
Microbial Detection and Measurement Technologies
Various methods are being applied or developed to detect microorganisms and measure their activities:
• Traditional recovery methods were discussed in the context of their application to mesophilic anaerobes to investigate a recall of infant formula that was presumptively implicated in a case of infant botulism in the United Kingdom. Eric Johnson of the University of Wisconsin’s Food Research Institute provided the presentation for Scott Donnelly of Wyeth Nutritionals, Burlington, Vt., who was unable to be present.
The recovery methods were used to (1) determine bioload of the raw material, (2) test air, water, ingredients, and product contact surfaces for Clostridium botulinum, and (3) propose specifications for mesophilic anaerobes. Existing methods used during the investigation included standard plate counts and established procedures delineated in the fourth edition of the Compendium of Methods for the Microbiological Examination of Foods (American Public Health Association, Washington, D.C.). In addition, a new method was developed specifically for this investigation.
Many of the media for mesophilic anaerobic sporeformers listed in the Compendium result in considerable variability and false positives, Johnson said. A major challenge that had to be overcome in the investigation was the lack of methods for enumerating spores in air. The formula manufacturer teamed up with Paddy O’Reilly of the University County Cork to develop a method involving modification of an SAS air sampler and gelatin and agar plates.
Another challenge that had to be overcome was the selection of a method for testing the water because an official, validated method does not exist. A filtration-based method described in the Official Journal of the European Communities was selected. Johnson’s review of the methods employed for the outbreak investigation showed the extraordinary measures to which a single case of botulism can lead and the considerable problems, even with traditional methods such as plating, that investigators can face.
• Measuring lag time, particularly from single spores of C. botulinum, and studying steps within the lag phase is where the biggest contribution can be made to better predicting growth in minimally heat processed (e.g., sous vide) foods. When a food safety problem involving minimally processed foods occurs, it will likely result from just a single spore, according to Mike Peck of the Institute of Food Research, Norwich, UK. If chances for microbial growth are low, as is often the case for C. botulinum, lag times and the resultant time to a 1-log increase can vary significantly, thus challenging food manufacturers’ abilities to ensure appropriate safety margins.
Peck described an image analysis system that he and colleagues developed for measuring time to lag steps (e.g., germination, emergence, time to one cell) and for quantifying the effect of stress conditions (pH, temperature, pretreatments) on lag. The system met several needs, including allowing identification of the same spore/cell at different developmental steps under automated, anaerobic, and temperature-controlled conditions.
Peck and colleagues concluded from their work to date that: (1) considerable variability exists in times to the different steps in the lag phase; (2) for individual spores there is poor correlation between times to the different lag steps; and (3) time to two cells cannot be predicted from time to germination or emergence. With further development of the system, Peck plans to examine the effect of environmental factors on each step in the lag phase, develop predictive models of the effect of food environment on each of the steps, establish which step is rate limiting, and establish why lag is so variable.
• Peptide nucleic acid (PNA)-based probes offer advantages over DNA-based based probes, particularly for hard-to-permeabilize microbial forms such as spores, encysted parasites, and Gram-positive bacteria. PNA is a pseudopeptide DNA mimic having an uncharged, achiral backbone. The unique chemical makeup of PNA confers a number of advantageous properties, including rapid hybridization kinetics, resistance to nucleases, and the ability to hybridize to positions on the ribosome that are inaccessible to DNA-based probes. PNA probes are also able to penetrate recalcitrant biological structures such as parasite cysts and Gram-positive bacterial cell walls. For this reason, PNA probes are especially useful with fluorescence in-situ hybridization (FISH), a rapid microbial identification method which uses fluorescently labeled nucleic acid probes to target variable regions of ribosomal RNA inside whole cells. Byron Brehm-Stecher of the University of Wisconsin-Madison’s Food Research Institute described the development of PNA-FISH probes and their combination with flow cytometry for the detection of Listeria spp.
--- PAGE BREAK ---
• Flow cytometry systems are composed of fluidic, optic, and electronic components. Flow cytometry involves laser-based irradiation of single cells containing bound fluorochromes; with the resultant light scattering providing cell size information. Systems allow rapid and quantitative analysis of individual cells. The technology was developed in late 1950 and is used primarily in clinical settings. Kristi Harkins of Advanced Analytical Technologies, Inc. (AATI), Ames, Iowa, described the design and applications of an analyzer, the RBD2100, developed by AATI and its successful application to B. subtilis spores and vegetative cells, Cryptosporidium parvum oocysts, Listeria spp., and Escherichia coli O157:H7.
• Fluidized bed capture uses molecules covalently bound via patented surface chemistry to glass beads to bind and hold target organisms and spores. Liquification of the sample before running it through the device at flow rates greater than 100 mL/min leads to fluidization of the beads and capture of microorganisms onto bead surfaces. The organisms are then detected via immunological or genetic testing. The procedure eliminates the need to pre-enrich a food sample and can be done in 30 min from the time of sample collection to final result. The technology may be able to detect as few as 100 cfu/mL, in an optimized format and depending on the capture ligand. This technology has been successfully used with Bacillus spores, E. coli O157, Listeria spp., and Salmonella.
Fluidized bed capture is the antithesis to flow cytometry, said Bart Weimer of Utah State University. Microbe detection with fluidized bed capture uses large volumes and enables alternatives to chemiluminescence. Weimer described the development of a number of rapid “capture technology” sensors—ImmunoFlow™, ImmunoDNA®, GlycoBind®, and TissueTag ®—for detecting spores and microbes in food and the environment.
• Multiplexing allows testing of many molecules simultaneously. Microarrays (spatially encoded micro-sized spots, typically on a slide) and microbeads (magnetic system of micro-sized encoded beads) are common multiplexing systems. Joydeep Lahiri of Corning, Corning, N.Y., provided insight into different types of multiplexing systems that Corning is working with as part of its research program aimed at development of advanced tools for biotechnology. He detailed state-of-the-art cDNA (long oligo), protein, and membrane protein and lipid microarrays.
Lahiri also described achievements made in their work to overcome issues associated with microbead assays. For example, to address the problem of the autofluorescence that is associated with current polymeric materials, Corning is using microbarcodes that are made of pristine glass, which is chemically inert, and contain rare earths, which emit only narrow fluorescence bands.
He also described work in assay detection systems ranging from label displacement via fluorescence resonance energy transfer (FRET) or quenching to “true” label-free detection (without fluorescent, radioactive, or other labels) via optical biosensors, electrochemistry, and mass spectroscopy. A microplate-based label independent detection system that he described is based on changes in refractive index and has a detection goal of about 1 × 10–6 refractive index units (molecular weight <500).
Corning’s applications of microarrays and microbeads build on the organization’s strengths in photonics. To date, applications have been in organic and biochemical technologies, rather than food.
• Micro/nano bioanalytical systems allow detection of pathogenic bacteria and viruses at the level of the single cell. Antje Baeumner of Cornell University described these systems and their advantages over traditional methods. Microanalytical systems offer speed (e.g., 10–15 min), sensitivity, reliability, inexpensive cost, simplicity for the user, portability, multi-analyte high-throughput capability, and new application areas. Based on specific optical RNA biosensors for E. coli and C. parvum, microbioanalytical systems are being developed for the Dengue virus and Bacillus anthracis; their detection is combined with sample preparation. The systems use the same biological principles as in simple biosensors; however, novel sample-preparation systems, novel molecular biology amplification (nucleic acid sequence–based amplification, NASBA) chambers, and intricate hybridization channel patterns are being developed.
The principles and techniques of the system’s modules are a laser-induced cell lysis system, interdigitated ultramicroelectrodes as transducers, NASBA amplification on the chip, and modeling and fabrication of optimized micromixers. Systems are specifically designed for RNA, basically providing amplification of mRNA as PCR does for DNA; however, the system does not produce false positives from dead microorganisms and does not require thermal cycling. Detection limits as low as femtomolar and single-cell range were demonstrated. System challenges include macroworld integration, analysis of appreciable volumes (from 1 mL to 100 L), and miniaturization of sample preparation.
--- PAGE BREAK ---
• The GeneExpert instrument platform developed by Cepheid, Sunnyvale, Calif., and described by Kurt Petersen performs all the steps, from sample preparation to final validated result, of DNA and RNA molecular detection technologies. The platform is being used in a U.S. Postal Service pilot program to detect biothreat agents. Other applications being developed include studying human infectious diseases, antibiotic resistance, foodborne pathogens, agricultural and veterinary diseases, and cancer.
The platform provides proper sample preparation, internal controls, simultaneous monitoring of multiple DNA targets, and a supply of single-dose reagents. It also overcomes the false-positive and false-negative problems associated with PCR, enabling an investigator to look for the needle in the haystack (e.g., one Listeria monocytogenes cell in a 300-g sample).
• Ganglioside-liposome nanovesicle immunoassay has been newly developed and shown to detect botulinum and cholera toxins at femto/attomolar concentrations. Described by Richard Durst of Cornell University, this nanovesicle immunoassay offers extremely good sensitivity and speed (15–20 min from the point of mixing the sample with liposome solution and test strip application). The assay can detect toxins in food and water with only moderate loss of sensitivity. Bioassays such as this use antigen–antibody interactions, nucleic acid hybridization, or natural receptor binding for recognition; liposomes or NASBA for signal enhancement; optical or electrochemical methods for detection; and flow-injection liposome analysis or migration strip, fluorometric tube, or microfluidic assays as formats. Gangliosides are useful in such bioassays because they can serve as toxin receptors (e.g., GM1 is the receptor ganglioside for the toxin produced by Vibrio cholera). Liposomes are useful because their composition can be controlled and tailored, their surface modified by conjugation or insertion, and their size and surface-tag concentration controlled; a variety of detection methods are possible; and lysis, when needed, gives instantaneous amplification.
Consensus on Research Needs
The outcomes of the discussions among the small groups were presented in a final plenary session that was immediately followed by development of consensus on research needs for addressing the unresolved questions and concerns in order to advance food quality and safety.
Three primary areas were identified as key for future investigations: (1) fundamental knowledge of spores, i.e., why spores are resistant and dormant; (2) sampling issues; and (3) microbial detection and measurement technology. Research in any of these areas will benefit the other areas. Furthermore, because many types of microorganisms could be considered difficult to measure, i.e., dormant, the conclusions of this forum may have broad application to mold and yeast spores, parasitic cysts, viruses, and VBNC microorganisms. A fourth item—the need to integrate the various pertinent disciplines (e.g., engineering, materials science, physical chemistry, microbiology) for a multidisciplinary approach—was also identified as important for future work.
• Fundamental Knowledge of Spores. There are extraordinary opportunities to ultimately understand dormancy and extreme resistance of bacterial spores and similar dormant microbial systems (e.g., VBNC microorganisms). Spore components (e.g., core, cortex, and inner membrane) must be studied at the architectural/molecular/material science level. Study at this level will contribute to the understanding of the basic mechanisms of spore resistance, dormancy, germination, and heterogeneity. Further, the outcome of such research will elucidate the physical state of the cortex and how its peptidoglycan acts at that level, clarify the status of the protoplast, enable understanding of the role of CaDPA in dormancy and resistance, and show how CaDPA and other agents (e.g., alkyl amines) act in germination.
Such information could lead to the development of specific targets for spore inhibition and detection. One participant said it would be great to be able to discover a generic component on the spore surface to which molecules could then be attached to induce superdormancy (thus completely inactivating a spore via a coating technique). Alternatively, with new information on how spores may be induced out of dormancy, the cells could be more easily inactivated.
--- PAGE BREAK ---
• Sampling/Capture Issues. As a reflection of the difficulty of overcoming sampling/capture issues, one participant said, “If we could solve the capture problem, detection would be a piece of cake.” Extracting or removing spores from their food matrices presents substantial challenges. However, results of some of the ongoing research offer the prospect that microorganisms may not always have to be extracted from the food matrix in the future. Although technologies allowing identification of microorganisms and their toxins continue to advance, sample preparation challenges remain because of the varied food matrices and, hence, different sample classes (e.g., grains, raw meat and poultry, spray-dried ingredients, powders, flavors). Furthermore, not only are mechanisms needed for overcoming the challenges presented by different food matrices, but tools or methods for validating the mechanisms are also needed.
• Microbial Detection and Measurement Technology. To conduct these much-needed investigations into fundamental spore physiology, nanotechnology and other contemporary techniques are crucial. As highlighted by several participants, considerable advances are rapidly being made in many areas. Techniques such as nanotechnology, PNA probes, flow cytometry, multiplexing, RNA biosensors, NASBA, and the GeneXpert platform hold tremendous potential for being effectively employed in this fundamental research. Why nanotechnology? Because genetic identification, automation, and use of lower concentrations of cells and materials can permit faster and more accurate, precise, and economical research into these dormant cell systems.
Dormant Microbes: Research Needs
These and other advanced techniques and systems will enable new insight into the fundamental aspects of spore physiology and dormant systems, and the understanding and detection of whole cells, cellular constituents, and metabolic products. With this insight, food manufacturers would then be better able to solve the challenges and problems associated with spores by being able to determine and apply practical, appropriate rapid “corrections” of processing parameters in various food matrixes/environments. Further, qualitative and quantitative nondestructive testing could be a great advantage for quality assurance systems, including off-line and in-line measurements that could be continuous.
The impressive capabilities of nano- and other advanced technologies raise several questions, however. What ultimate lower limits of detection will be attainable and meaningful? What will be the ultimate roles and advantages of these techniques in food safety and quality, whether in the field or the laboratory? Can these advanced techniques improve our understanding of dose response? Where will all the opportunities lie in food safety epidemiology? What are the many roles these advanced techniques might play in intentional contamination of food or the food environment?
One participant pointed out that nanotechnology will not be “the holy grail,” however. He said application of nanotechnology will bring about many more issues for research, which should be addressed at the outset by the food industry directing, via funding, specific development of biosensors with the food sector in mind. Elaborating, he said that nanotechnology presents pitfalls when trying to apply it to particulate-containing samples, as is the case of food. Thus, work in surface chemistry, definition of surface interactions, and basic microbiology is needed to effectively use the technology for food matrices.
Although the advancing work in functional genomics was not a focus of the meeting, it is understood that genomics and expression arrays also hold considerable promise for fundamental research and detection of dormant organisms.
--- PAGE BREAK ---
• Disciplinary Integration/Multidisciplinary Approach. Ongoing and new technological developments must move from basic to applied research and meld together to effect enhanced food quality, safety, and public health. Thus, different disciplines having a bearing on or potentially contributing to the effective connection of basic and applied research must be integrated for a multidisciplinary approach to research. Further cross-disciplinary education and training will contribute immensely to the positive impact and outcome of basic and applied research.
• Research Benefits. Who needs the outcomes of the research? Who will benefit from application of resources to the areas described above? Regulatory agencies could use this knowledge for process validation, foodborne disease surveillance, risk assessment, criteria for standards, and outbreak investigations. Homeland Security and Defense could use the information and findings in military situations as well as in terrorism control, effective anti-terrorism measures, and general quality assurance. The food industry could obviously benefit from programs aimed at food safety, spoilage control, and quality management related to bacterial spores, yeast spores, mold spores, and cysts. New research and development opportunities, such as sporeforming probiotics and new preservation techniques could result as well. Environmental control efforts also could benefit from this research, to design enhanced integrated pest management systems and to help monitor and control contamination of air, hospitals, public buildings, and strategic facilities by these resistant spores. In fact, these research outcomes could prove fundamental for many biological systems other than spores and other dormant microorganisms.
Thus, the findings in this area will greatly impact not only the economics of the food industry but also public health and public confidence in the food supply.
Research Summit Participants
ORGANIZER
Frank F. Busta, Professor Emeritus and Emeritus Head, Dept. of Food Science and Nutrition, Univ. of Minnesota, St. Paul.
PARTICIPANTS
Arjan J. van Asselt, NIZO Food Research, Ede, The Netherlands; Antje J. Baeumner, Cornell Univ., Ithaca, N.Y.; Robert Beelman, Pennsylvania State Univ., University Park; Dane Bernard, Keystone Foods, LLC, West Conshohocken, Pa.; M.A.J.S. “Tiny” van Boekel, Wageningen Univ., The Netherlands; Byron Brehm-Stecher, Univ. of Wisconsin-Madison; Pilar De Massaguer, State University of Campinas, Brazil; Richard Durst, Cornell University, Ithaca, N.Y.; John Floros, Pennsylvania State Univ., University Park, Grahame Gould, formerly of Unilever Research, Bedford, UK; John Hanlin, General Mills, Inc., Minneapolis, Minn.; Kristi R. Harkins, Advanced Analytical Technologies, Inc., Ames, Iowa; Kai Lai Grace Ho, Praxair, Inc., Burr Ridge, Ill.; Eric Johnson, Univ. of Wisconsin-Madison; Stephen Knabel, Pennsylvania State Univ., University Park; Joydeep Lahiri, Corning, Inc., Corning, N.Y.; James Lindsay, U.S. Dept. of Agriculture, Beltsville, Md.; William Lulves, Minute Maid Co., Apopka, Fla.; Arthur Miller, Food and Drug Administration, College Park, Md.; Michael Peck, Institute of Food Research, Norwich, UK; Kurt Petersen, Cepheid, Sunnyvale, Calif.; Rukma Reddy, Food and Drug Administration, Summit-Argo, Ill.; Tom Romick, ConAgra Foods, Irvine, Calif.; Peter Setlow, Univ. of Connecticut, Farmington; Guy Skinner, Food and Drug Administration, Summit-Argo, Ill.; John Sofos, Colorado State Univ., Fort Collins; Pieter ter Steeg, Unilever, Vlaardingen, The Netherlands; Cindy Stewart, CSIRO Food Science Australia, North Ryde; Bart Weimer, Utah State Univ., Logan; Ahmed Yousef, Ohio State Univ., Columbus; Guopeng (John) Zhang, Canadian Inovatech, Inc., Abbotsford, British Columbia.
PROGRAM COMMITTEE
Larry Beuchat, Univ. of Georgia, Griffin; Kathryn Boor, Cornell Univ., Ithaca, N.Y.; Martin Cole, CSIRO Food Science Australia, North Ryde; Michael Davidson, Univ. of Tennessee, Knoxville; Grahame Gould, formerly of Unilever Research, Bedford, UK; Dennis Heldman, Rutgers Univ., New Brunswick; Dallas Hoover, Univ. of Delaware, Newark; Eric Johnson, Univ. of Wisconsin-Madison; Theodore Labuza, Univ. of Minnesota, St. Paul; Peter McClure, Unilever, Bedford, UK; Virginia Scott, National Food Processors Association, Washington, D.C.; Peter Setlow, Univ. of Connecticut, Farmington; Charles Sizer, National Center for Food Safety and Technology, Summit-Argo, Ill.; Cynthia Stewart, CSIRO Food Science Australia, North Ryde.
by Rosetta Newsome
Director, IFT Dept. of Science and Communications
