The Prevalence, Transmission, and Control of C. difficile in Food
FOOD SAFETY AND QUALITY
Formerly known as Clostridium difficile, Clostridioides difficile is one of the most common microorganisms responsible for gastrointestinal illnesses in hospitals and nursing homes worldwide. It is a gram-positive spore-forming, anaerobic, human pathogen. It produces three toxins: toxin A, toxin B, and CDT. C. difficile PCR ribotype 027 has been well studied compared to others; however, other C. difficile strains (including ribotypes 0126, 018, 056, 078, and 244) have been linked to severe C. difficile infection (CDI) outbreaks with high relapse rate and mortality. The transmission of C. difficile continues to be a challenge for healthcare professionals and food microbiologists since CDI and mortality from it have been on the rise. This organism is commonly found in the environment and thus occurs in a variety of animal-based and plantbased foods, indicating its likely transmission as a foodborne pathogen.
C. difficile Infection and Epidemiology
C. difficile colonizes human intestines, which are a major source of this pathogen. Adult humans (up to 17.5%) and infants/neonates (up to 37%) are asymptomatic carriers of C. difficile (Scähffler and Breitrück 2018). CDI causes a broad spectrum of clinical illness including diarrhea, colitis, pseudomembranous colitis, and fulminant colitis. The current model that describes the development of CDI proposes that antibiotic-associated disruption of the structure and/or function of the human intestinal microbiota enables colonization of C. difficile. The colonized organism causes CDI in the absence of a functional immune response (Ollech et al. 2016). CDI has also been linked to the following susceptible populations: the very young, the elderly, the immunocompromised, and patients with underlying illnesses such as inflammatory bowel diseases, ulcerative colitis, and Chron’s disease.
Over the past two decades, the incidence of CDI has significantly increased worldwide. The U.S. Centers for Disease Control and Prevention has declared CDI an urgent threat. According to the 2016– 2017 European point prevalence study, C. difficile ranked sixth among microorganisms responsible for healthcare-associated infections in Europe (Suetens et al. 2018). In many other countries of the world, CDI presents a significant problem to patients and healthcare facilities due to its morbidity and mortality, long hospital stays, and extensive costs (Hensgens et al. 2013; Barbut et al. 2017). However, CDI is no longer limited to hospitals and nursing homes and is becoming more prevalent in the community. More than 25% of all CDI cases are estimated to be community- acquired. Interestingly, these cases appear to affect groups not previously at risk, including younger populations and those without antibiotic exposure.
Diagnosis and Detection
The toxigenic culture method is the most dependable method for CDI diagnosis; this is a two-step method: First, C. difficile cells are isolated from stool samples on a selective microbiological media. Second, in vitro toxin production by C. difficile isolates is examined. Additional methods, such as enzyme immunoassays for toxins, glutamate dehydrogenase assay, nucleic acid amplifications tests (polymerase chain reaction, loop-mediated isothermal amplification, helicasedependent amplification assay, and microarray technologies), and ultrasensitive assays that use single molecule array technology, can also be used as diagnosis and detection tools (Guery et al. 2019). Culture methods that include non-selective enrichment followed by selective microbiological media are used for isolation of C. difficile from food (Candel-Pérez et al. 2019).
C. difficile in the Agricultural Environment
A ubiquitous organism in nature, C. difficile has been isolated from soils, saltwater and freshwater sources, wastewater, animal manure, and food animals (Candel-Pérez et al. 2019). In food animals, C. difficile has been found in their intestinal tracts, which means it can be distributed via fecal contamination of soils, water, food processing environments, and final products. The organism has been extensively described in both healthy pigs and pigs with diarrhea, with spore and toxin detection ranging from 1.4% to 96% in piglets with normal feces and ranging from 23% to 93% in diarrheagenic piglets. The reported prevalence of C. difficile in cattle varies from 0% to 60% depending on the geographical location. C. difficile has been linked to diarrhea in calves and dairy cows. A limited number of studies have focused on C. difficile in poultry. Available data indicate that the prevalence of C. difficile in food animals decreases with age and that C. difficile colonization has been detected whether or not the animals have signs of illness (Rodriguez et al. 2016). Whole genome sequencing has been used to study the genetics of C. difficile and the epidemiology of CDI. Knetch et al.(2014) reported that farmers and pigs were colonized with identical C. difficile clones, supporting the interspecies transmission hypothesis between animals and humans.
C. difficile in Foods
The presence of C. difficile endospores in final food products can be explained by initial contamination of raw materials and/or ingredients, cross-contamination in the food processing plants, and unintentional production of endospores during food processing. C. difficile spores have been detected in different types of food products, including seafood, vegetables, and meats with a prevalence ranging from 2.9% to 66.7% (Rodriguez et al. 2016). These prevalence figures stress the potential risk of CDI associated with consuming raw and under-processed foods. C. difficile was reported in vacuum-packaged ground meat (Bouttier et al. 2010) and chilled vacuum- packed meats with blown pack spoilage (Broda et al. 1996). Its regular association with meat animals combined with its routine isolation from meats provide strong evidence for the likely involvement of C. difficile as a foodborne illness agent.
Because animal manure can contain spores of C. difficile, the application of composted manure to land is a possible risk of C. difficile transfer to the food chain. Irrigation (or washing) with C. difficile contaminated water can result in contamination of vegetables and fruits with the organism (Rupnik and Sanger 2010). C. difficile has been isolated from raw vegetables, including cucumbers, onions, radishes, carrots, mushrooms, and ready-to-eat salads. A 2008 study of ready-to-eat salads mostly imported from European Union countries and sold in Glasgow, Scotland, detected C. difficile in three of 40 salad samples (Lund and Peck 2015).
Inactivation of C. difficile spores by thermal treatment has been assessed, and cooking food at a minimum of 96°C for 15 minutes produced an inhibitory effect on C. difficile spores (Rodriguez- Palacios et al. 2011). Minimally processed fruits and vegetables are treated below these temperatures. Consequently, fruits and vegetables grown with organic fertilizer containing C. difficile spores and/or irrigation water or washed with contaminated water could be potential vehicles of CDI (Rodriguez-Palacios et al. 2013).
CDI Prevention & Probiotics
Treatment of CDI can involve either one or more antibiotics (e.g., metronidazole, vancomycin, fidaxomicin) or the transplantation of fecal microbiota. However, substantial efforts to find alternative treatments for the prevention/decline of CDI are underway. Some of these efforts have led to the development and use of a nonabsorbable rifamycin antibiotic, microbiota-based drugs, human monoclonal antibodies targeting toxins A and B, and two vaccines (Guery et al. 2019). Perhaps the most interesting emerging way to treat C. difficile is the use of probiotics.
The Food and Agriculture Organization and World Health Organization define probiotics as “live microorganisms which when administered in adequate amounts confer a health benefit to the host.” Probiotics have been shown to have a beneficial effect in prevention of antibiotic-associated diarrhea, treatment of acute infectious diarrhea as well as CDI. Even though the role of probiotics in the prevention of CDI has been controversial, a mounting body of evidence from in vitro, preclinical, and clinical studies suggests that probiotics play a role in the primary prevention of CDI. Several physiological mechanisms for protective and beneficial effects of probiotics on C. difficile colonization resistance have been suggested, including antimicrobial activity, intestinal barrier protection, immunomodulation, and alteration of intestinal microbiota (Ollech et al. 2016).
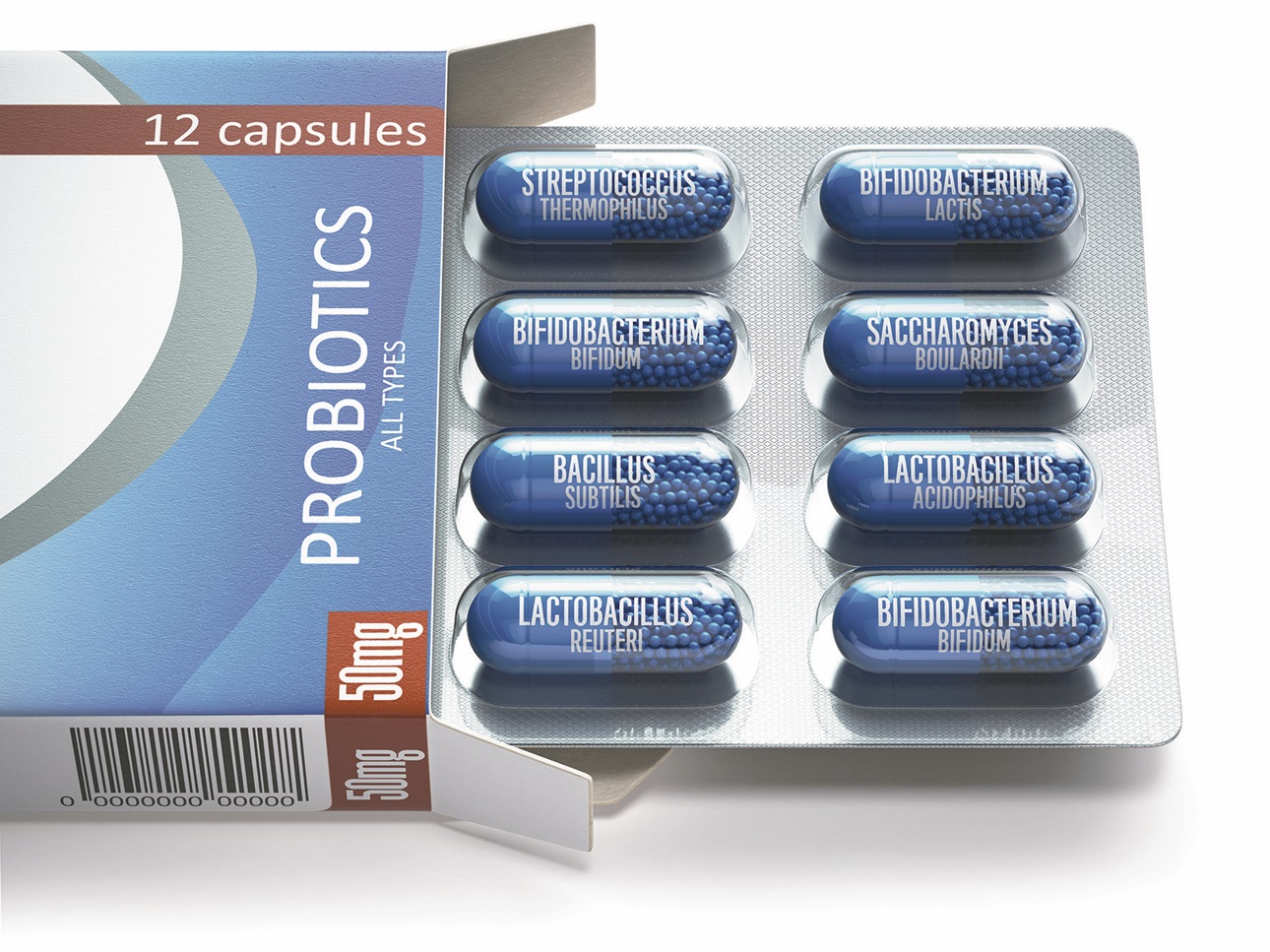
Three commercial probiotics were clinically tested against C. difficile. They are Bio-K, VSL#3 and HOWARU Restore. Bio-K is comprised of three strains of Lactobacillus (Lactobacillus acidophilus CL1285, Lactobacillus casei LBC80R, and Lactobacillus rhamnosus CLR2). A phase 3 trial (randomized and double-blinded) demonstrated that Bio-K use during antibiotic treatment was linked to lower incidence of antibiotic- or C. difficile associated diarrhea (Gao et al. 2010).
VSL#3 is a probiotic preparation of eight live freeze-dried bacterial species. These species are normal residents of the human gastrointestinal microflora, including four strains of lactobacilli (Lactobacillus casei, L. plantarum, L. acidophilus, and L. delbrueckii subsp. bulgaricus), three strains of bifidobacteria (Bifidobacterium longum, B. breve, and B. infantis) and Streptococcus salivarius subsp. thermophilus. When VSL#3 was tested in a multicenter randomized double-blind study in patients exposed to antibiotic course, the results demonstrated a decrease in cases of antibiotic-associated diarrhea, but no CDI cases were reported (Selinger et al. 2013).
HOWARU Restore has four organisms: Lactobacillus acidophilus NCFM, Lactobacillus paracasei Lpc-37, Bifidobacterium animalis subsp. lactis Bl-04, and B. lactis Bi-07. HOWARU Restore at different dosages was assessed in a randomized clinical trial. The results from the trial demonstrated a decrease in CDI in the probiotic group (Ouwehand et al. 2014).
Even though the scientific community has known about C. difficile for decades, transmission of the organism through food has not been well-documented. The role of food animals and food in C. difficile carriage and CDI must be further studied to fill the knowledge gap. The prospective work is expected to be challenging but also rewarding, especially considering the serious nature of CDI in susceptible populations and the medical costs associated with it.
REFERENCES
Barbut, F., S. Bouée, L. Longepierre, et al. 2017. “Excess Mortality Between 2007 and 2014 Among Patients With Clostridium difficile Infection: a French Health Insurance Database Analysis.” J. Hosp. Infect. 98: 21–28.
Bouttier, S., M. C. Bare, B. Felix, et al. 2010. “Clostridium difficile in Ground Meat, France.” Emerg. Infect. Dis. 16: 733–735.
Candel-Pérez, C., G. Ros-Breeuezo, C. Martínez-Graciá. 2019. “A review of Clostridioides [Clostridium] difficile Through the Food Chain.” Food Microbiol. 77: 118–120.
Gao, X. W., M. Mubasher, C. Y. Fang, et al. 2010. “Dose-Response Efficacy of a Proprietary Probiotic Formula of Lactobacillus CL1285 and Lactobacillus casei LBC80R for Antibiotic Associated Diarrhea Prophylaxis in Adult Patients.” Am. J. Gastroenterol. 105: 1636–1641.
Guery, B., T. Galperine, F. Barbut. 2019. “Clostridioides difficile: Diagnosis and Treatments.” BMJ2019:366:14609. doi: 10.1136/bmi.14609.
Hensgens, M. P. M., A. Goorhuis, O. M. Dekkers, et al. 2013. “All-Cause Disease- Specific Mortality in Hospitalized Patients With Clostridium difficile Infection: a Multicenter Cohort Study.” Clin. Infect. Dis. 56: 1108–1116.
Knetch, C. W., T. R. Connor, A. Mutreja, et al. 2014. “Whole Genome Sequencing Reveals Potential Spread of Clostridium difficile Between Humans and Farm Animals in the Netherlands, 2002 to 2011.” Euro Surveill 19: 20954.
Lund, B.M., and M. W. Peck. 2015. “A Possible Route for Foodborne Transmission of Clostridium difficile?” Foodb. Path. Dis.12(3): 177–182.
Ollech, J. E., N. T. Shen, C. V. Crawford, et al. 2016. “Use of Probiotics in Prevention and Treatment of Patients With Clostridium difficile Infection. Best Pract. Res. Clin. Gastroenterol. 30: 111–118.
Ouwehand A. C., C. DongLian, X. Weijan, et al. 2014. “Probiotics Reduce Symptoms of Antibiotic Use in a Hospital Setting: a Randomized Dose Response Study. Vaccine 32: 458–463.
Rodriguez, C., B. Taminiau, J. Van Broeck, et al. 2016. “Clostridium difficile in Food and Animals: A Comprehensive Review.” Adv. Exp. Med. Biol. 4: 65–92.
Rodriguez-Palacios, A., S. Borgmann, T. R. Kline. 2013. “Clostridium difficile in Foods and Animals: History and Measures to Reduce Exposure.” Anim. Health Res. Rev. 14: 11–12.
Rodriguez-Palacios, A., M. Koohmaraie, J. T. Lejeune. 2011. “Prevalence, Enumeration, and Antimicrobial Agent Resistance of C. difficile in Cattle at Harvest in the United States.” J. Food Prot. 74: 1618–1624.
Rupnik, M. and J. G. Sanger. 2010. “Clostridium difficile: Its Potential as a Source of Foodborne Illness.” Adv. Food Nutr. Res. 60: 53–66.
Selinger, C. P., A. Bell, A. Cairns, et al. 2013. “Probiotic VSL#3 Prevents Antibiotic- Associated Diarrhea in a Double-Blind, Randomized, Placebo-Controlled Clinical Trial.” J. Hosp. Infect. 84: 150–165.
Suetens, C., K. Latour, T. Kärki, et al. 2018. “Prevalence of Healthcare-Associated Infections Estimated Incidence and Composite Antimicrobial Resistance in Acute Care Hospitals and Long-Term Care Facilities: Results From Two European Point Prevalence Surveys, 2016 to 2017.” Euro Surveill 23: e1002150.